3870
Parallel Proton and Deuterium Metabolic Imaging1Yale University, New Haven, CT, United States
Synopsis
Keywords: Deuterium, Deuterium, DMI
1H MRSI and DMI can provide crucial metabolic information to facilitate evaluations of neurological diseases. In this work, we applied interleaved 1H/2H methodology to achieve time-efficient, parallel 1H MRSI and DMI acquisition. We first describe the necessary sequence adjustments for both nuclei, including the re-optimization of the VAPOR water suppression scheme and the incorporation of a 2H equilibrium pulse. Phantoms were then used to demonstrate that high quality 1H MRSI and DMI data can be acquired in parallel. The interleaved 1H MRSI-DMI sequence is compatible with other interleaved MRI-DMI sequences and can be implemented for a complete MRI-DMI protocol.
Introduction
1H MRS(I) is a powerful, non-invasive tool to spatially map metabolic profiles, thereby providing unique and complementary information to that provided by MRI. It has been shown to effectively facilitate the evaluation of various neurological diseases1 and has been emerging from a research method to a clinical imaging modality.1H MRSI maps a static metabolic profile, representing the metabolic pool sizes without information on dynamic turnover. Broader metabolic characterization to also provide dynamic metabolism information and can be obtained with isotope tracer methods, typically based on13C, HP13C and most recently 2H MRSI (DMI).Acquiring 1H MRSI and DMI in addition to conventional, anatomical MRI can benefit clinical evaluations of neurological diseases. However, a sequential acquisition of 1H MRSI and DMI in addition to a clinical MRI session is too time consuming. Previously, we have developed a strategy to achieve parallel 1H-2H acquisitions using multi-nuclear interleaved technology3, and have applied it to a variety of MRI sequences without prolonging the original MRI scan time4. In this work, we apply a similar method to incorporate DMI into 1H MRSI scans. We present our first interleaved 1H MRSI-DMI sequence and demonstrate its ability to obtain parallel 1H MRSI and DMI data from a metabolite phantom.
Methods
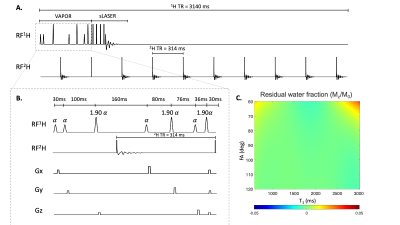
Parallel 1H MRSI-DMI pulse sequence. 1H MRSI was achieved with semi-LASER (localization by adiabatic spin-echo refocusing) localization (TR/TE = 3140/144 ms) and VAPOR water suppression. The excitation pulse (Shinnar-Le Roux, Tp = 2ms, BW = 3045Hz) selects a 10 mm axial slice after which wideband uniform rate and smooth truncation (WURST) modulated gradient-modulated offset independent adiabaticity (GOIA) pulses (Tp = 4 ms, BW = 11.5 KHz )5 were used for in-plane selection.2H pulse-acquisitions necessary for 3D DMI were placed in the delays periods of the 1H MRSI RF pulse sequence. To maintain a constant 2H TR, DMI sequence elements also need to be inserted during the VAPOR module, entailing a re-optimization of the VAPOR pulse angles and delays. The customized VAPOR scheme allows one full DMI block and one 2H excitation pulse to be inserted after the 3rd and the 7th water suppression pulse, fulfilling the requirements of both nuclei. For one 1H TR (3140 ms), nine 2H pulse acquisitions and one 2H equilibrium pulse were inserted (Figure 2A).
Phantom Measurements. All data were acquired on a Bruker 4T 94cm Medspec system (Bruker Corporation. Ettlingen, Germany) using a single channel TEM 1H volume coil and a 4 channel phased array 2H coil. Phantoms containing creatine (10 mM), DMSO (0-5 mM), D2O (0.5%) and DMSO-D6 (0.2%-0.5%) in agar were measured using the interleaved MRSI-DMI sequence. 1H MRSI (21x19 matrix over 210x190 mm2, circular k-space encoding) was acquired in parallel with DMI (13x9x11 matrix over 260x180x220 mm3, spherical k-space encoding). Non-water-suppressed 1H MRSI was acquired to perform phase correction on the same phantom. The referencing MRI was acquired on the axial plane using a GRE sequence.
Results
Figiure. 1B shows the VAPOR water suppression sequence optimized to allow the insertion of one full 2H pulse-acquire block and one 2H equilibrium pulse. An average residual water fraction (Mz/M0) of -0.0003±0.003 over the range of simulated nutation angle and T1 (Figure 1C) was comparable to that of the original VAPOR method and led to adequate water suppression in all studies.Figure 2 shows the results on phantoms using the newly developed 1H/2H pulse sequence. Example 1H spectra corresponding to the highlighted pixels in Figure 2A (red boxes) accurately depicted the composition of both phantoms with high spectral quality (Figure 2B). A relatively homogeneous DMSO and creatine map (Figure 2D-E) was obtained. DMSO/creatine (Figure 2F) accurately reflects the ratio between two compounds correctly. 2H spectra of the highlighted regions in Figure 2A (green boxes) are shown in Figure 2C. The DMSO/D2O ratio measured between two peaks appeared to be uniform and was consistent with composition of the metabolite mixture (Figure 2C).
Discussions
We successfully designed a parallel 1H MRSI-DMI sequence by interleaving 2H pulse-acquire blocks into the delays inherently provided by the 1H MRSI method. The VAPOR water suppression pulse angles and delays were re-optimized to enable 2H sequence elements insertion while maintaining a constant TR and providing a sufficient acquisition window for DMI. 1H MRSI scans often employ long TRs, and therefore are ideal candidates in interleaved DMI pulse acquisitions. High quality 1H MRSI and DMI were obtained on phantoms, indicating a high level of water suppression and the capacity of parallel 1H and 2H MR data acquisition using the sequence. In addition to being used alone, the interleaved 1H MRSI-DMI sequence can also be used with other interleaved MRI-DMI sequences, such as FLAIR-DMI3 and become part of a complete MRI-DMI protocol4.Acknowledgements
This research was funded, in part, by NIH grant NIBIB R01-EB025840.References
1. Öz G, Alger JR, Barker PB, et al. Clinical Proton MR Spectroscopy in Central Nervous System Disorders. Radiology. 2014;270(3):658-679.2. De Feyter HM, Behar KL, Corbin ZA, et al. Deuterium metabolic imaging (DMI) for MRI-based 3D mapping of metabolism in vivo. Science Advances. 2018;4(8):eaat7314.3. Liu Y, De Feyter HM, Fulbright RK, McIntyre S, Nixon TW, de Graaf RA. Interleaved fluid-attenuated inversion recovery (FLAIR) MRI and deuterium metabolic imaging (DMI) on human brain in vivo. Magn Reson Med. 2022.4. Liu Y, De Feyter HM, Fulbright RK, McIntyre S, Nixon TW, de Graaf RA. A Multi-Sequence, Interleaved MRI-DMI Protocol for Human Brain. 2022.5. Kumaragamage C, Coppoli A, Brown PB, et al. Short symmetric and highly selective asymmetric first and second order gradient modulated offset independent adiabaticity (GOIA) pulses for applications in clinical MRS and MRSI. Journal of Magnetic Resonance. 2022;341:107247.
Figures

1H MRSI acquired using the interleaved sequence on agar phantoms containing creatine (1mM) DMSO (0-5mM), D2O (0.5%) and DMSO-D6 (0.2-0.5%). (B-C) Show examples of localized 1H and 2H spectra in pixels indicated in (A). The DMSO and creatine map shown in (D-E) are calculated by integrating the area under the corresponding peaks and normalizing them to water signal obtained in the non-water-suppressed reference scan. (F-G) illustrate the DMSO/creatine and DMSO-D6/D2O obtained from 1H MRSI and DMI respectively.