3860
Neurochemical differences between 1p/19q codeleted and non-codeleted gliomas assessed by in vivo magnetic resonance spectroscopy1Sorbonne University, UMR S1127, INSERM U1127, CNRS UMR 7225, Paris Brain Institute - ICM, Paris, France, 2Department of Radiology, Neuroradiology Unit, ASST Spedali Civili University Hospital, Brescia, Italy, 3Center for Magnetic Resonance Research, Department of Radiology, University of Minnesota, Minneapolis, MN, United States, 4Pathology Unit, Department of Molecular and Translational Medicine, University of Brescia, Brescia, Italy, 5Laboratory R Escourolle, University Hospital La Pitié Salpêtrière, Paris, France, 6Department of neuroradiology, University Hospital La Pitié Salpêtrière, Paris, France, 7Department of neurology 2, University Hospital La Pitié Salpêtrière, Paris, France, 8Center for Neuroimaging Research (CENIR), ICM, Paris, France
Synopsis
Keywords: Spectroscopy, Cancer
Mutations in the genes encoding for isocitrate dehydrogenase (IDH), and 1p/19q codeletion are genetic alterations that can be often found in low-grade gliomas and are associated with better prognosis and response to treatment. Nevertheless, the biological effects of the 1p/19q codeletion are still not well known.
In this study, we compared the neurochemical profile of IDH-mutated, 1p/19q codeleted gliomas and their non-codeleted counterparts using in vivo magnetic resonance spectroscopy. We showed that the only metabolite that differed significantly between the two groups was cystathionine, pointing to this metabolite as the most useful biomarker for the identification of 1p/19q codeleted gliomas.
Introduction
Diffuse gliomas are the most common primary brain tumors, whose diagnostic and prognostic stratification relies today on the status of few molecular markers1. These include mutations in the genes encoding for isocitrate dehydrogenase 1 or 2 (IDH1/2)2, and complete codeletion of the chromosome arms 1p and 19q (1p/19q codeletion)3, the latter being found exclusively in IDH-mutated gliomas and conferring the best prognosis4. Molecular aberrations induce metabolic changes in cancer cells, whose consequences on tumorigenesis, tumor progression and response to treatment are not well known5. Specifically, IDH mutations cause the abnormal accumulation of D-2-hydroxyglutarate (2HG)6, an established marker of these mutations. While several studies addressed metabolic differences in IDH-mutated vs. IDH-wild-type gliomas7, little has been reported so far on altered metabolism in 1p/19q codeleted compared to non-codeleted gliomas.The aim of this study was to compare the in vivo neurochemical profile between IDH-mutated, 1p/19q codeleted gliomas and their non-codeleted counterparts using proton magnetic resonance spectroscopy (MRS).
Methods
Human subjects. Thirty-one subjects with IDH1-mutated gliomas (13 1p/19q codeleted and 18 1p/19q non-codeleted) were retrospectively included in the study based on the availability a PRESS acquisition before any treatment including surgery.MR acquisitions. Acquisitions were performed at 3 T. 3D FLAIR images were acquired to position the spectroscopic volume of interest (VOI) in the glioma. MR spectra were acquired using a single-voxel PRESS sequence optimized for 2HG detection8 (TR = 2.5 s, TE = 97 ms, 128 averages). Unsuppressed water scans were acquired from the same VOI for metabolite quantification and eddy current corrections using the same parameters as water suppressed spectra.
Post-processing. Single-shots were frequency and phase aligned in Matlab using the total choline signal at 3.22 ppm. Spectral analysis was performed using LCModel and a simulated basis set. Metabolite quantification was performed using the water signal as a reference and correcting for water and metabolite longitudinal and transverse relaxation constants. Only metabolites with Cramér-Rao lower bounds (CRLB) < 50% for more than half of the subjects, in at least one of the two groups of codeleted or non-codeleted subjects, were considered for further analysis. A two-tailed t-test was used to compare metabolite concentrations between groups, accounting for multiple comparisons.
Results
Examples of spectra acquired in IDH1-mutated, 1p/19q codeleted and non-codeleted gliomas are shown in Figure 1. The two groups did not show a significant difference in the mean total creatine linewidth, which was 4.5 ± 1.0 Hz (range 3.6 – 7.3 Hz) and 5.2 ± 1.1 Hz (range 3.6 – 8.0 Hz) in codeleted and non-codeleted gliomas, respectively. Nineteen metabolites were included in the statistical analysis: 2HG, ascorbate, aspartate, betaine, citrate, choline compounds, cystathionine, GABA, glutamate, glutamine, glutathione, myo-inositol, N-acetyl aspartate + N-acetyl-aspartyl-glutamate, lactate, scyllo-inositol, serine, succinate, taurine, and total creatine. We observed a significantly higher cystathionine concentration in codeleted vs. non codeleted gliomas (mean ± standard deviation: 2.33 ± 0.98 mM vs. 0.93 ± 0.94 mM, p = 0.0004, Figure 2). Median CRLBs associated with cystathionine quantification were 11% and 33% in two groups, respectively.Cystathionine was detected with concentration > 1.4 mM and CRLB < 25% in all 1p/19q codeleted gliomas except one. In these tumors, the cystathionine signal can be visually identified at 2.7 ppm, where it partially overlaps with the aspartate signal (Figure 1A). In the non-codeleted group, cystathionine concentration was < 1 mM in 11 out of 18 subjects, while it ranged from 1 to 2.9 mM in the remaining subjects. Using an arbitrary threshold of 1 mM concentration or 25% CRLB, the sensitivity and specificity of this method for 1p/19q codeletion identification were 92% and 61%, respectively.
Other metabolite concentrations did not differ significantly between groups.
Discussion and conclusions
Our results are in agreement with a previous study showing significantly higher cystathionine levels in codeleted vs. non-codeleted gliomas from both ex vivo glioma tissue analysis and in vivo edited MRS9. Edited MRS allows to reliably detect cystathionine thanks to the lack of overlap between the cystathionine signal at 2.7 ppm and other resonances10. However, edited sequences and advanced post-processing tools are not yet always available in clinical settings, which limits the use of edited MRS for diagnostic and prognostic purposes in the clinic. In contrast, PRESS is a standard MRS method that could be more easily employed in the clinical practice.Cystathionine was the only metabolite showing a significant difference in concentration between codeleted and non-codeleted gliomas, highlighting the role of this neurochemical as a potentially powerful biomarker in 1p/19q codeleted gliomas.
Our results suggest that the addition of optimized MRS to current multi-modal clinical protocols may significantly increase the sensitivity and specificity to determine both IDH mutational status and 1p/19q codeletion via quantification of 2HG and cystathionine, respectively.
Further studies will aim at understanding the biological mechanisms underlying the abnormal accumulation of cystathionine also in a subset of non-codeleted gliomas and to evaluate the clinical relevance of this metabolite for the assessment of treatment response and progression.
Acknowledgements
The authors would like to thank Edward J. Auerbach, Ph.D. for implementing MRS sequences on the Siemens platform. FBr and SL acknowledge support from Investissements d’avenir [grant number ANR-10-IAIHU-06 and ANR-11-INBS-0006]. FBr acknowledges support from Agence Nationale de la Recherche [grant number ANR-20-CE17-0002-01]. DD and MM acknowledge support from following National Institutes of Health (NIH) grants: BTRC P41 EB015894 and P30 NS076408. MM acknowledges support from the NIH grant U01CA269110. MS acknowledges support from INCa-DGOS-Inserm_12560 (SiRIC CURAMUS).References
1. Kristensen BW, Priesterbach-Ackley LP, Petersen JK, Wesseling P. Molecular pathology of tumors of the central nervous system. Ann Oncol. 2019;30(8):1265-1278. doi:10.1093/annonc/mdz164
2. Yan H, Parsons DW, Jin G, et al. IDH1 and IDH2 mutations in gliomas. N Eng J Med. 2009;360(8):765-773.
3. Reifenberger J, Liu L, James CD, Wechsler W. Molecular genetic analysis of oligodendroglial tumors shows preferential allelic deletions on 19q and 1p. 1994;145(5):16.
4. Cairncross JG, Zlatescu MC, Lisle DK, et al. Specific genetic predictors of chemotherapeutic response and survival in patients with anaplastic oligodendrogliomas. J Nat Canc Inst. 1998;90(19):7.
5. Hvinden IC, Cadoux-Hudson T, Schofield CJ, McCullagh JSO. Metabolic adaptations in cancers expressing isocitrate dehydrogenase mutations. Cell Rep Med. 2021;2(12):100469.doi:10.1016/j.xcrm.2021.100469
6. Dang L, White DW, Gross S, et al. Cancer-associated IDH1 mutations produce 2-hydroxyglutarate. Nature. 2009;462(7274):739-744. doi:10.1038/nature08617
7. Branzoli F, Marjańska M. Magnetic resonance spectroscopy of isocitrate dehydrogenase mutated gliomas: current knowledge on the neurochemical profile. Curr Opin Neurol. 2020;33(4):413-421. doi:10.1097/WCO.0000000000000833
8. Choi C, Ganji SK, DeBerardinis RJ, et al. 2-hydroxyglutarate detection by magnetic resonance spectroscopy in IDH-mutated patients with gliomas. Nat Med. 2012;18(4):624-629. doi:10.1038/nm.2682
9. Branzoli F, Pontoizeau C, Tchara L, et al. Cystathionine as a marker for 1p/19q codeleted gliomas by in vivo magnetic resonance spectroscopy. Neuro Oncol. 2019;21(6):765-774. doi:10.1093/neuonc/noz031
10. Branzoli F, Deelchand DK, Sanson M, Lehéricy S, Marjańska M. In vivo 1 H MRS detection of cystathionine in human brain tumors. Magn Reson Med. 2019;82(4):1259-1265. doi:10.1002/mrm.27810
Figures
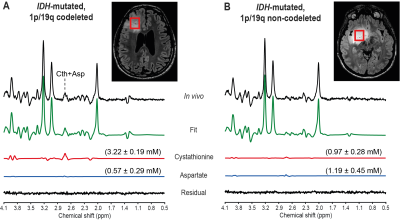
Figure 1. In vivo optimized PRESS spectra in IDH1-mutated, 1p/19q codeleted and non-codeleted gliomas. In vivo optimized PRESS spectra acquired at 3 T in two subjects with (A) an IDH1-mutated, 1p/19q codeleted glioma and (B) an IDH1-mutated, 1p/19q non-codeleted glioma are shown together with the LCModel fit, the cystathionine and aspartate contributions and the residual. Shown in parenthesis is the metabolite concentration ± CRLB (mM). No line-broadening was applied. The VOIs are shown on axial 3D FLAIR images.
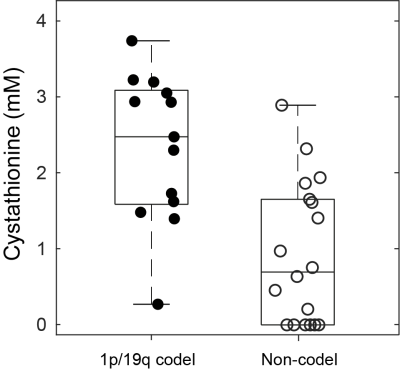
Figure 2. Cystathionine concentration in 1p/19q codeleted vs. non-codeleted gliomas. Box-plots of cystathionine concentrations in 1p/19q codeleted (filled circles) vs. non-codeleted gliomas (empty circles). For each box, the central mark indicates the median concentration, and the bottom and top edges indicate the 25th and 75th percentiles, respectively. Circles represent values from individual subjects.