3777
Lifelong collaborative learning improves the performance of complex muscle MR image segmentation tasks
Francesco Santini1,2, Jakob Wasserthal2, Abramo Agosti3, Xeni Deligianni1,2, Kevin R Keene4, Hermien E Kan5, Stefan Sommer6,7,8, Christoph Stuprich9, Fengdan Wang10, Claudia Weidensteiner1,11, Giulia Manco12, Valentina Mazzoli13, Arjun Desai14, and Anna Pichiecchio12,15
1Basel Muscle MRI, Department of Biomedical Engineering, University of Basel, Basel, Switzerland, 2Research Coordination Team, Department of Radiology, University Hospital Basel, Basel, Switzerland, 3Department of Mathematics, University of Pavia, Pavia, Italy, 4Department of Neurology, Leiden University Medical Center, Leiden, Netherlands, 5C.J. Gorter MRI Centre, Department of Radiology, Leiden University Medical Center, Leiden, Netherlands, 6Siemens Healthineers International AG, Zurich, Switzerland, 7Swiss Center for Musculoskeletal Imaging (SCMI), Balgrist Campus, Zurich, Switzerland, 8Advanced Clinical Imaging Technology (ACIT), Siemens Healthineers International AG, Lausanne, Switzerland, 9University Hospital Erlangen, Erlangen, Germany, 10Peking Union Medical College, Beijing, China, 11Radiological Physics, Department of Radiology, University Hospital Basel, Basel, Switzerland, 12Advanced Imaging and Radiomics Center, IRCCS Mondino Foundation, Pavia, Italy, 13Department of Radiology, Stanford University, Stanford, CA, United States, 14Departments of Electrical Engineering & Radiology, Stanford University, Stanford, CA, United States, 15Department of Brain and Behavioural Sciences, University of Pavia, Pavia, Italy
1Basel Muscle MRI, Department of Biomedical Engineering, University of Basel, Basel, Switzerland, 2Research Coordination Team, Department of Radiology, University Hospital Basel, Basel, Switzerland, 3Department of Mathematics, University of Pavia, Pavia, Italy, 4Department of Neurology, Leiden University Medical Center, Leiden, Netherlands, 5C.J. Gorter MRI Centre, Department of Radiology, Leiden University Medical Center, Leiden, Netherlands, 6Siemens Healthineers International AG, Zurich, Switzerland, 7Swiss Center for Musculoskeletal Imaging (SCMI), Balgrist Campus, Zurich, Switzerland, 8Advanced Clinical Imaging Technology (ACIT), Siemens Healthineers International AG, Lausanne, Switzerland, 9University Hospital Erlangen, Erlangen, Germany, 10Peking Union Medical College, Beijing, China, 11Radiological Physics, Department of Radiology, University Hospital Basel, Basel, Switzerland, 12Advanced Imaging and Radiomics Center, IRCCS Mondino Foundation, Pavia, Italy, 13Department of Radiology, Stanford University, Stanford, CA, United States, 14Departments of Electrical Engineering & Radiology, Stanford University, Stanford, CA, United States, 15Department of Brain and Behavioural Sciences, University of Pavia, Pavia, Italy
Synopsis
Keywords: Software Tools, Machine Learning/Artificial Intelligence
An open-source, federated-learning-based segmentation software termed Dafne (Deep Anatomical Federated Network) is presented. This software continuously adapts the deep learning models used for the segmentation (currently for the muscles of the leg and thigh) based on the input of the users, who are in multiple institutions. This software was validated through data usage statistics of more than 50 users and through a retrospective study on 38 datasets of patients with suspected myositis, showing that the continuous learning approach is able to improve and generalize the performance of the original models.Introduction
Deep learning (DL) algorithms are commonly used for segmenting MR images. These algorithms learn from a set of training data and are then able to generalize the application to real-world images. For these algorithms to work, the training data needs to be sufficiently large and representative of the type of images encountered during real-life application, which is more challenging than other imaging modalities because of the variety of protocols and contrasts. This is particularly true for skeletal muscle MRI, because of the deformable geometry, the natural variation across subjects, and the presentation of different pathologies, most of which are rare.In this work, we present and validate a system termed Dafne (Deep Anatomical Federated Network) that implements federated learning by distributing the segmentation software, complete with user interface, to multiple users, and the model is updated after each user’s usage. In this aspect, this model implements distributed lifelong learning. With this approach, the models can be trained on data from multiple institutions and multiple diseases while preserving data privacy. This approach is validated by collecting performance data from users and on a controlled set of datasets with suspected myositis.
Methods
Dafne was released in 2021, and it has a client/server architecture. Both the client and the server are developed in Python, and are released as free software under a Gnu General Public License (GPLv3)1,2. The user interface allows image visualization and is responsible for downloading the deep-learning models from the server and performing the segmentation. After automatic segmentation, the user has the possibility to correct and refine the automatic segmentation with a set of editing tools (mask and contour editing mode, registration- and interpolation-based mask propagation, edge snapping, and more). The model is then updated by performing an incremental learning step on the client side on the data that is therefore never transmitted outside the user’s institution. The refined model is finally transmitted to the server, where it is then validated and merged with the base version (Fig. 1).Two deep-learning models are currently provided, based on a modified V-Net architecture3, for the segmentation of the muscles of the thigh and the leg.
For the validation of the system, dice similarity indices (DSI) between the automatic segmentation and the refined masks were transmitted from every client at every usage. No restriction was posed on the contrast or acquisition protocol used as input data.
As a controlled validation, 38 T1-weighted anonymized datasets containing acquisitions of the leg were retrospectively retrieved from the PACS archive of one of the sites containing patients with suspected myositis. Of these datasets, 25 were segmented using the Dafne workflow by two separate readers (12 by one reader and 13 by the other), and the refined models incrementally merged with the base model (incremental training). After segmenting the remaining 13 datasets, the incremental training phase was used to validate the models by comparing their performance with the manual segmentation phase. For each dataset, we recorded the difference between the DSI performance of the subsequent models and the DSI performance before the incremental training phase. Two linear regression models were fitted on the time course of the differences in DSI to establish whether the learning was effective.
Results
Dafne currently has more than 50 users from multiple institutions. During the period in which data were recorded for this abstract, the median DSI from clients was 0.80 over 256 valid data points. Sample automatic segmentations produced by one site during systematic usage by multiple users, showing improvement in the segmentation, is shown in Figure 2.The validation phase resulted in 13 merge events for the 25 incremental training datasets (the client only transmits the updated model when the network is available so that each merge event might contain the refinements from multiple datasets). On average, the DSI improved both in the incremental training and validation sets. From the linear fitting, the DSI improved by 0.009 per event (p < 0.001, 95% confidence interval 0.008-0.010) for the incremental training data, and by 0.007 per event (p < 0.001, 95% C.I. 0.006-0.008). The plots are shown in Fig. 3. The code and the data to produce these results are available online4 and are released under an Apache v2.0 free license.
Discussion
In this work, we demonstrated that a lifelong learning approach as implemented in Dafne is effective for the segmentation of medical images, and it can generalize to new data, provided that the incremental training includes data with similar characteristics. Although we retain the characteristics of data privacy and distributed learning, our approach differs from traditional federated learning in that the model is trained incrementally rather than in batches. As with other lifelong learning cases, the performance of the model changes over time and can also decay on older datasets if the new inputs have different characteristics. Implementing Dafne as a coherent, user-interface-based solution is of crucial importance for the continuous evolution of the model and to ensure human oversight when the models are applied to a vastly variable set of input data.Acknowledgements
No acknowledgement found.References
1. Dafne. https://www.dafne.network/. Accessed October 25, 2022.
2. dafne-imaging. GitHub. https://github.com/dafne-imaging. Accessed November 6, 2022.
3. Agosti A, Shaqiri E, Paoletti M, et al. Deep learning for automatic segmentation of thigh and leg muscles. Magma N. Y. N 2021 doi: 10.1007/s10334-021-00967-4.
4. GitHub - dafne-imaging/dafne-evaluation: Evaluation and figure generation for Dafne. https://github.com/dafne-imaging/dafne-evaluation. Accessed November 6, 2022.
Figures
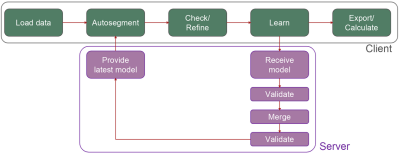
Fig 1: Dafne workflow schematics, with separation between client and server tasks
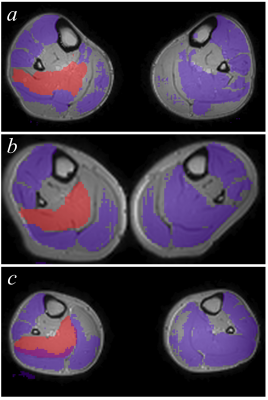
Fig 2: Sample segmentation performance on three subsequent days of usage for one site. The Soleus muscle (currently active muscle in the segmentation interface) is highlighted in red.

Fig 3: Evolution of the differential Dice Similarity Indices (DSI) on incremental training (left) and validation (right) datasets during the validation phase. The differential DSIs are calculated as a difference between the DSI at each time point and the DSI at t=0. The thick red line represents the average differential DSI, and each time point represents a model merge event.
DOI: https://doi.org/10.58530/2023/3777