3775
HYBRID MULTI-LEVEL GRAPH NEURAL NETWORK FOR CARDIAC MAGNETIC RESONANCE SEGMENTATION1Harbin Institute of Technology, Harbin, China
Synopsis
Keywords: Data Processing, Segmentation
Due to the inherent locality of convolutional operations, convolution neural network (CNN) often exhibits limitations in explicitly modeling long-distance dependencies. In this paper, we propose a novel hybrid multi-level graph neural (HMGN) network that combines the CNN and graph neural network to capture both local and non-local image features at multiple scales. With the proposed patch graph attention module, the HMGN network can capture image features over a large receptive field, resulting in more accurate segmentation of cardiac structures. Experiments on two public datasets show the proposed method obtains improved segmentation performance over the state-of-the-art methods.Synopsis
Keywords: cardiac MR images, segmentation, graph neural network, multiple scalesMedical image segmentation methods typically utilize convolution neural network (CNN) with pooling layers to expand the receptive field for capturing high-level features. However, due to the inherent locality of convolutional operations, CNN often exhibits limitations in explicitly modeling long-distance dependencies. In this paper, we propose a novel hybrid multilevel graph neural (HMGN) network to segment cardiac magnetic resonance (MR) images that combines the CNN and graph neural network (GNN) to capture both local and non-local features at multiple scales. The HMGN network can capture image features over a large receptive field, resulting in more accurate segmentation of cardiac structures.
Introduction
Cardiovascular disease has become the leading cause of death worldwide according to the world health organization report[1]. In practice, doctors usually evaluate the patient heart function according to the functional parameters, the calculation of which relies on the cardiac MR image segmentation results. The target of medical image segmentation is assigning labels to pixels with specific semantic information, including tumors and organs, etc. With the development of deep learning, convolution neural network (CNN) has been widely applied in image segmentation. Tran et al.[2] proposed that applied fully convolutional networks (FCNs) to segment the cardiac MR image for efficient end-to-end training. Liu et al.[3] proposed that applied the U-Net model to exploit both low-level and high-level image features for multi-sequence cardiac MR image segmentation and achieved significant results. However, CNN pays more attention to local information and has limited ability to extract long-distance features. In contrast, graph neural network (GNN) is not restricted to fixed local connections and can establish flexible connections between nodes. Recently, many researchers dedicated to applying GNN in image segmentation. Lu et al.[4] proposed the Graph-FCN method which built a graph network based on the high-level feature map of FCN and defined a graph loss function to enhance the FCN feature extraction capability. Camillo et al.[5] proposed a joint GNN-CNN model to segment the multimodal MR image with brain tumors, converting the image segmentation problem to the graph node classification problem. However, nodes of the GNN-CNN model were obtained by the simple linear iterative cluster (SLIC)[6] algorithm which may take lots of the running time, as described in [7].Methods
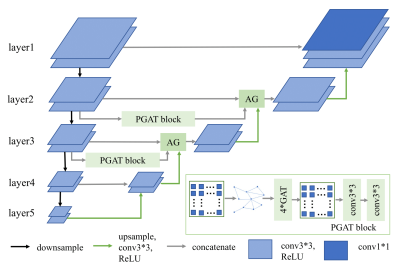
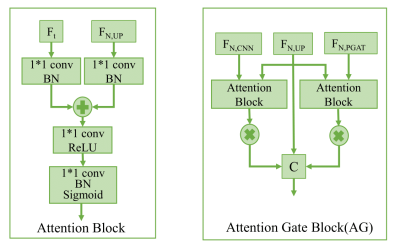
Fig.1 is the diagram of the proposed method. We first use the CNN encoder to extract image local features at multiple scales. To capture non-local features, we add the proposed patch graph attention (PGAT) block at layer 2 and layer 3. The PGAT block contains a graph generator, four graph attention (GAT)[8] blocks, a graph reprojector, and two 3∗3 convolutions. Specifically, we first divide the CNN feature map into patches and treat each patch as a node. The edges and adjacency matrix are obtained from the connections of nodes. Then we convert the feature map to the graph structure, and we calculate the average of the feature patch region corresponding to each node as the node initial features. The graph data is fed into cascaded multi-head GAT blocks to aggregate the neighbor node features. The output is reprojected to the corresponding patch region and concatenates two 3∗3 convolutions as post-processing. Furthermore, we add an attention gate[9] block on the skip connection paths to suppress irrelevant background regions as shown in Fig.2. The proposed method is implemented on the Pytorch framework using an RTX 3060-type GPU.Results and discussion
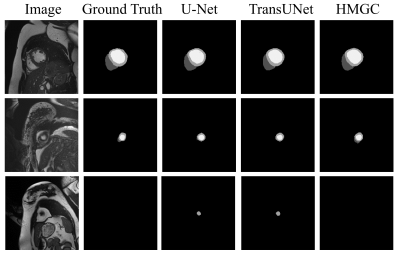
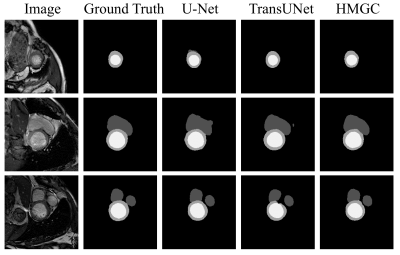
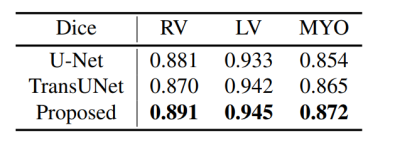
We use two datasets to validate the performance of the proposed method: Automatic Cardiac Diagnostic Challenge Medical Image Computing and Computer Assisted Intervention (ACDC-MICCAI) dataset and Multi-sequence Cardiac MR Segmentation Challenge (MS-CMRSeg) dataset. ACDC dataset was collected with an SSFP sequence along the short axis with slice thicknesses ranging from 5 to 8 mm. ACDC dataset contains 150 cine MR images, but only 100 have ground truth labels. We divided the labeled data into training data, validating data, and testing data with the ratio of 7:1:2. MS-CMRSeg dataset provides 45 cine MR images acquired under multi-sequence, namely the LGE, T2, and bSSFP. We select the bSSFP data as training and testing data. To avoid overfitting, we utilize augmentation strategies on the training data, including random rotating, flipping, zooming, and random cropping. The performance of the proposed HMGN method is compared with U-Net[10] and TransUNet[11]. Fig.3 and Fig.4 show the segmentation results of ACDC data and MS-CMRSeg data, respectively. Here we select three slices of each data for presentation. Table.1 present the quantitative comparison of different methods on ACDC datasets. Experimental results demonstrate that the proposed method can significantly improve the segmentation accuracy of RV, LV, and MYO in cardiac MR images compared with state-of-the-art methods.Conclusion
In this paper, we propose the HMGN model for cardiac MR image segmentation. Specifically, we utilize the proposed PGAT block to capture image features over a large receptive field. In addition, we combine CNN to obtain local features at different scales. We validate the performance of the proposed method on the ACDC dataset and MS-CMRSeg dataset. The segmentation results and quantitative analysis have demonstrated that the proposed method can achieve more accurate and reasonable segmentation results compared with U-Net and TransUNet even if the target boundary is illegible.Acknowledgements
No acknowledgement found.References
1. World Health Organization et al., “World health statistics 2020,” 2020.
2. Phi Vu Tran, “A fully convolutional neural network for cardiac segmentation in short-axis mri,” arXiv preprint arXiv:1604.00494, 2016.
3. Yashu Liu, Wei Wang, and et al., “An automatic cardiac segmentation framework based on multi-sequence mr image,” in International Workshop on Statistical Atlases and Computational Models of the Heart. Springer, 2019, pp. 220–227.
4. Yi Lu, Yaran Chen, and et al., “Graph-fcn for image semantic segmentation,” in international symposium on neural networks. Springer, 2019, pp. 97-105.
5. Camillo Saueressig, Adam Berkley, and et al., “A joint graph and image convolution network for automatic brain tumor segmentation,” in International MICCAI Brainlesion Workshop. Springer, 2022, pp. 356–365.
6. Achanta, Radhakrishna, and et al. "SLIC superpixels compared to state-of-the-art superpixel methods." IEEE transactions on pattern analysis and machine intelligence 34.11 (2012): 2274-2282.
7. Jun Zhang, Zhiyuan Hua, and et al., “Joint fully convolutional and graph convolutional networks for weakly-supervised segmentation of pathology images,” Medical image analysis, vol. 73, pp. 102183, 2021.
8. Petar Velickovic, Guillem Cucurull, and et al., “Graph attention networks,” arXiv preprint arXiv:1710.10903, 2017.
9. Ozan Oktay, Jo Schlemper, and et al., “Attention u-net: Learning where to look for the pancreas,” arXiv preprint arXiv:1804.03999, 2018.
10. Olaf Ronneberger, Philipp Fischer, and et al., “U-net: Convolutional networks for biomedical image segmentation,” in International Conference on Medical image computing and computer-assisted intervention. Springer, 2015, pp. 234–241.
11. Jieneng Chen, Yongyi Lu, and et al., “Transunet: Transformers make strong encoders for medical image segmentation,” arXiv preprint arXiv:2102.04306, 2021.
Figures