3771
Deep learning-based auto-segmentation of neck nodal metastases on longitudinal MR images using self-distilled masked image transformer1Medical Physics, Memorial Sloan Kettering Cancer Center, New York, NY, United States, 2Radiation Oncology, Memorial Sloan Kettering Cancer Center, New York, NY, United States, 3Radiology, Memorial Sloan Kettering Cancer Center, New York, NY, United States
Synopsis
Keywords: Segmentation, Cancer
Manual segmentation of normal and tumor tissues on MRI is a traditional approach that is still used, but it is a very challenging and time-consuming method requiring a high level of precision and has shown inter-reader contouring variability. Therefore, semi- or fully automated segmentation algorithms are essential to segment tumors such as neck nodal metastases. The present study aimed to apply the previously developed deep learning-based self-distilled masked image transformer method for auto-segmenting neck nodal metastases on longitudinal T2-weighted MR images.
Introduction
Manual segmentation of normal and tumor tissues on MRI is a traditional approach that is still used, but it is a very challenging and time-consuming method requiring a high level of precision and has shown inter-reader contouring variability1. Therefore, semi- or fully automated segmentation algorithms are essential to segment tumors such as neck nodal metastases2. Most head and neck cancer patients present with neck nodal metastases at the time of diagnosis. At present, we manually delineate nodes to measure tumor volumes and extract quantitative imaging biomarkers from the region of interest (ROI) to help identify MRI-based criteria for approaches such as dose de-escalation in radiation therapy3. Self-supervised learning (SSL) has recently demonstrated success in medical image segmentation using deep learning convolutional networks4. We recently developed an SSL deep learning (DL) method called Self-distillation learning with Masked Image modeling using vision Transformers (SMIT) for 3D multi-organ segmentation from CT and MRI4. This method was trained to learn the anatomical context of the underlying images using masked image modeling as a pre-text task with 3,643 imaging examinations (i.e., 602,708 images) obtained from cancer patients4. The present study is the first to build on this previously developed DL-based SMIT method and apply it for auto-segmentation of neck nodal metastases on longitudinal T2 weighted MR images.Methods
Patient: Our institutional review board approved this prospective dose de-escalation clinical trial. Written informed consent was obtained from all eligible Human papillomavirus-related HPV (+) oropharyngeal squamous cell carcinoma (OPSCC) patients with neck nodal metastases before enrollment5. Between February 2018 and December 2020, 123 HPV+ OPSCC patients were enrolled in the MRI study and were eligible for the 30Gy dose de-escalation trial. Longitudinal MRIs were performed at pretreatment and weekly (1-3 weeks) during chemo-radiation treatment (CRT) for the OPSCC patients.MRI data Acquisition: MRI scans were performed on a Philips 3T scanner (Ingenia; Philips Healthcare, Netherlands) using a neurovascular phased-array coil. The standard MR acquisition comprised multi-planar T2-weighted (T2w) (repetition time [TR] = 4000 ms, echo time [TE] = 80 ms, number of averages [NA] = 2, and number of slices [NS] = 40, matrix = 256 × 256, slice thickness = 5 mm, the field of view [FOV] = 20–24 cm) and precontrast and post-contrast T1w imaging (TR = 600 ms, TE = 8 ms, NA = 2, NS = 40, slice thickness = 5.0 mm; matrix = 256 × 256, FOV = 20–24 cm). The total MRI acquisition time was approximately 30 min.
Region of Interest Contouring: A team of two radiation oncologists with four years of experience and one neuroradiologist with more than ten years of experience manually contoured the neck nodal metastases on longitudinal T2w images using ITK- SNAP, in consensus6. The manually contoured regions of interest (ROI) were used as references (ground truth) for DL-based auto segmentation. The tumor volume was extracted from the manually delineated ROIs using ITK- SNAP.
Deep learning-based auto-segmentation method: We applied the DL-based SSL-SMIT method4 to auto-segment the neck nodal metastases on longitudinal T2w MRI images. We pretrained the 3D Swin-Unet segmentation model using SMIT and finetuned it with the Swin-Unet model. A total of 461 T2w MRI datasets from 123 patients were used in this study, among which 93, 10, and 20 patient datasets were used for training, validation, and testing, respectively. We used a patch size of 128×128×128 for training. During testing, the same patch size, 128×128×128, with a sliding window of 0.5, was implemented to segment the whole image volume. The segmented results were then resampled to the original voxel size to provide the auto segmentation for the neck nodal metastases. Fine-tuning was performed using five-fold cross-validation. The tumor volume was extracted from the auto-segmented ROIs using ITK- SNAP.
Statistical analysis: Segmentation accuracy was evaluated using the Dice similarity coefficient (DSC), with manual delineation serving as the ground truth. Statistical differences in the tumor volumes extracted from the manual and DL-based auto-segmented ROIs were computed using R software (Wilcoxon rank-sum test). P-values < 0.05 were considered statistically significant.
Results
The DL-based SMIT method achieved a median DSC of 0.89 on pretreatment MR images, 0.89 on week 1, 0.82 on week 2, and 0.79 on week 3 MR images during the chemo-radiation therapy for patients on 30Gy dose de-escalation trial. Figure 1 shows two representative segmentation cases with both manual and auto segmentation of the ROIs from pretreatment to week 3 during CRT. The statistical distribution of segmentation accuracy (DSC) is shown in Figure 2. The mean value of the manually contoured tumor volume at pretreatment was 8.38±7.01 cm3. In contrast, the SSL SMIT-based pretreatment volume was 8.40±7.29 cm3, and there is no significant difference between the manual contour and SMIT-based auto-segmented volumes (P=0.48) at pretreatment. Figure 3 shows two representative cases of longitudinal contoured volumes with SMIT and manual delineation for OPSCC patients during CRT.Discussion and Conclusion
We successfully applied our DL-based SMIT method for auto-segmenting neck nodal metastases on longitudinal T2-w MR images. Our proposed model is potentially valuable for improving the efficiency of head and neck cancer treatment planning and is less time-consuming.Acknowledgements
This work was supported by the NIH grants NIH U01 CA211205 (ASD), R01 CA238392-01A1 (NYL), and NIH/NCI Cancer Center Support Grant P30 CA008748.References
1. Despotović I, Goossens B, Philips W. MRI segmentation of the human brain: challenges, methods, and applications. Comput Math Methods Med. 2015;2015:450341. doi:10.1155/2015/450341
2. Schouten JPE, Noteboom S, Martens RM, et al. Automatic segmentation of head and neck primary tumors on MRI using a multi-view CNN. Cancer Imaging. 2022/01/15 2022;22(1):8. doi:10.1186/s40644-022-00445-7
3. Riaz N, Sherman E, Pei X, et al. Precision Radiotherapy: Reduction in Radiation for Oropharyngeal Cancer in the 30 ROC Trial. JNCI: Journal of the National Cancer Institute. 2021;113(6):742-751. doi:10.1093/jnci/djaa184
4. Jiang, J., Tyagi, N., Tringale, K., Crane, C., Veeraraghavan, H. Self-supervised 3D Anatomy Segmentation Using Self-distilled Masked Image Transformer (SMIT). In: Medical Image Computing and Computer Assisted Intervention, MICCAI 2022. vol 13434. pp 556–566. doi: 10.1007/978-3-031-16440-8_53
5. Paudyal R, Oh JH, Riaz N, et al. Intravoxel incoherent motion diffusion-weighted MRI during chemoradiation therapy to characterize and monitor treatment response in human papillomavirus head and neck squamous cell carcinoma. Journal of magnetic resonance imaging: JMRI. Apr 2017;45(4):1013-1023. doi:10.1002/jmri.25523
6. Yushkevich PA, Yang G, Gerig G. ITK-SNAP: An interactive tool for semi-automatic segmentation of multi-modality biomedical images. Annu Int Conf IEEE Eng Med Biol Soc. Aug 2016;2016:3342-3345. doi:10.1109/embc.2016.7591443
Figures
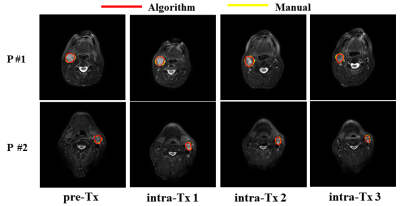
Figure 1 MR images from two representative patients showing manual and auto-segmented neck nodal metastases. The red and yellow contours indicate the manual and algorithm (auto) segmentation, respectively.
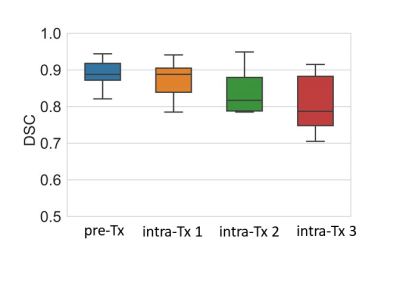
Figure 2 The box plot exhibits DSC accuracy for neck nodal metastases segmentation pre and post-treatment.
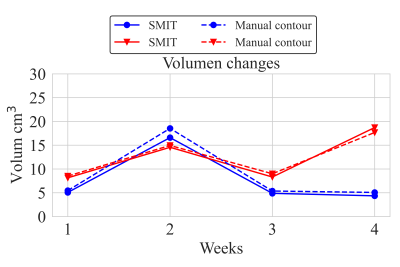
Figure 3 The neck model metastases volumes obtained with DL-based SMIT and manual segmentation for two representative patients pre and post-treatment.