3767
Complementarity-aware multi-parametric MR image feature fusion for abdominal multi-organ segmentation1Paul C. Lauterbur Research Center for Biomedical Imaging, Shenzhen Institutes of Advanced Technology, Chinese Academy of Sciences, Shenzhen, China, 2Guangdong Provincial Key Laboratory of Artificial Intelligence in Medical Image Analysis and Application, Guangzhou, China, 3University of Chinese Academy of Sciences, Beijing, China, 4Peng Cheng Laboratory, Shenzhen, China
Synopsis
Keywords: Segmentation, Body
T1-weighted in-phase and opposed-phase gradient-echo imaging is a routine component in abdominal MR imaging. Organ segmentation with the acquired images plays an important role in identifying various diseases and making treatment plans. Despite the promising performance achieved by existing deep learning models, further investigation is still needed to effectively exploit the information provided by different imaging parameters. Here, we propose a complementarity-aware multi-parametric MR image feature fusion network to extract and fuse the information of paired in-phase and opposed-phase MR images for enhanced abdominal multi-organ segmentation. Extensive experiments are conducted, and better results are achieved when compared to existing methods.Introduction
Abdominal multi-organ segmentation is essential for many clinical applications, including disease diagnosis (e.g., liver failure) and treatment planning (e.g., computer-aided surgery) 1. Automatic segmentation algorithms are needed to help physicians achieve fast and accurate image interpretation. The majority of existing studies in this field focused on analyzing CT images 2–5. Recently, MR imaging has been increasingly employed in clinical practice. There are studies showing that MR imaging is more accurate than CT for the diagnosis of hepatic, adrenal, and pancreatic diseases 6, and T1-weighted dual gradient-echo in-phase and opposed-phase MR imaging is a valuable tool for the diagnosis of a variety of benign and malignant processes in the abdomen 7,8. Therefore, abdominal multi-organ segmentation based on the acquired multi-parametric MR images (paired in-phase and opposed-phase MR images) is meaningful and necessary.Deep learning-based multi-parametric MR image segmentation has been extensively investigated for brain and prostate segmentation 9–11. There are three major types of deep learning-based image feature fusion: early fusion 12-14, late fusion15,16, and multi-layer fusion 11,17,18. However, how to effectively extract and fuse the information obtained from images acquired with different imaging parameters is still an open question.
In this study, we propose a complementarity-aware multi-parametric MR image feature fusion network. Particularly, we develop an enhanced baseline model by using residual connections and the attention mechanism and propose a complementarity-aware feature fusion method to fuse the features of different modalities. Extensive experiments are conducted by utilizing the MR images provided by the CHAOS challenge 1. Promising results are generated.
Methodology
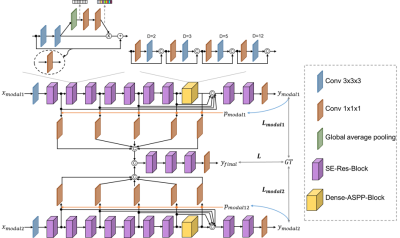
The architecture of our proposed framework is depicted in Fig. 1. Two network streams are designed to extract information from the paired in-phase and opposed-phase MR images. The basic building block of our network is SE-Res-Block, which combines the residual connections with the channel-wise attention module to increase the network’s effectiveness in image information extraction. To achieve accurate segmentation of both large and small organs, pooling operations are not utilized, and instead, a series of dilated convolutions of different dilation rates are introduced to increase the field-of-view (FOV) of the network. Then, multi-level image features are fused to make the final predictions.A complementarity-aware multi-parameter image feature fusion module is proposed to fully exploit the complementary information. Specifically, probability maps ($$$p_{modal1}$$$) and ($$$p_{modal2}$$$) in Fig. 1) are generated independently from the predictions of the two modalities. Supposing $$$y_{modal1}$$$ is the predicted class classification probability of modality 1, $$$p_{modal1}$$$ (same for $$$p_{modal2}$$$) is calculated as $$$p_{modal1}=1-\frac{\prod \limits_{i=1}^N y_{modal1,i}}{{(1/N)}^N}$$$, where N is the number of classes and $$$y_{modal1,i}$$$ is the predicted probability map of class i. Each value in $$$p_{modal1}$$$ or $$$p_{modal2}$$$ can indicate the certainty of the network’s prediction of the voxel in the 3D MR image. Therefore, they can indirectly represent the information entropy of the images. We utilize $$$p_{modal1}$$$ and $$$p_{modal2}$$$ to achieve our complementarity-aware image feature fusion by multiplying the multi-level image features with the corresponding certainty maps. Then, these features are fused by concatenation after one adaptation layer consists of a $$$1\times1\times1$$$ convolution operation. Final predictions ($$$y_{final}$$$) are made with the fused image features.
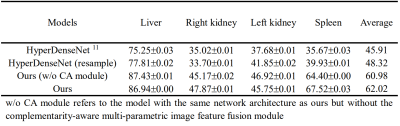
Two baseline models are implemented. HyperDenseNet is a recently published state-of-the-art multi-parametric MR image segmentation model 11. Another baseline has the same architecture as that of our proposed model but the multi-level features are fused directly after the adaptation layers without the complementarity-aware module. Experiments are conducted with the CHAOS challenge dataset 1. The challenge provides 20 images with labels for the segmentation of the liver, kidneys, and spleen. These 20 images are divided randomly into a training set of 12 images, a validation set of 2 images, and a test set of 6 images. Image patches of $$$32\times32\times32$$$ are extracted with a step of $$$14\times14\times14$$$ from each 3D image. We further propose a data resample strategy to balance the four classes during model training. Each experiment is replicated 3 times. No post-processing strategies are applied. Results are presented with $$$mean \pm s.d.$$$.
Results and Discussion
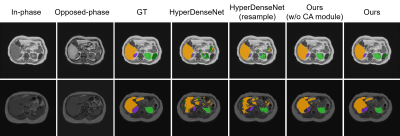
To quantitatively evaluate the segmentation performance, Dice similarity coefficient (DSC) is calculated and reported. Results are listed in Table 1. It can be observed that HyperDenseNet achieves the lowest scores among the different methods. Our proposed data resample strategy (HyperDenseNet (resample)) can slightly improve the segmentation accuracy. The proposed baseline network (Ours (w/o CA module)) can largely improve the segmentation score (12% absolute increase on the average DSC value), demonstrating the importance of information extraction. Our final model embracing the complementarity-aware module can achieve further enhanced segmentation performance, especially for the right kidney and the spleen. Therefore, both information extraction and fusion are crucial for the final segmentation performance. Example segmentation results are plotted in Fig. 2. It can be observed our proposed method can achieve better segmentation results compared to the comparison methods. Nevertheless, there are still misclassifications that should be improved in future studies.Conclusion
In this study, a complementarity-aware multi-parametric MR image feature fusion network is proposed for abdominal multi-organ segmentation. Experimental results demonstrate that the proposed method is able to enhance the segmentation accuracy of both large and small organs. The proposed method can be very helpful in clinical applications where multi-parametric MR imaging is widely adopted.Acknowledgements
This research was partly supported by Scientific and Technical Innovation 2030-"New Generation Artificial Intelligence" Project (2020AAA0104100, 2020AAA0104105), the National Natural Science Foundation of China (61871371, 62222118, U22A2040), Guangdong Provincial Key Laboratory of Artificial Intelligence in Medical Image Analysis and Application (2022B1212010011), the Basic Research Program of Shenzhen (JCYJ20180507182400762), Shenzhen Science and Technology Program (Grant No. RCYX20210706092104034), and Youth Innovation Promotion Association Program of Chinese Academy of Sciences (2019351).References
[1] Kavur, A. E. et al. CHAOS Challenge - combined (CT-MR) healthy abdominal organ segmentation. Med. Image Anal. 69, 101950 (2021).
[2] Gibson, E. et al. Automatic multi-organ segmentation on abdominal CT with dense V-networks. IEEE Trans. Med. Imaging 37, 1822–1834 (2018).
[3] Luo, X. et al. WORD: A large scale dataset, benchmark and clinical applicable study for abdominal organ segmentation from CT image. Med. Image Anal. 82, 102642 (2022).
[4] Ma, J. et al. Fast and low-GPU-memory abdomen CT organ segmentation: The FLARE challenge. Med. Image Anal. 82, 102616 (2022).
[5] Wang, Y. et al. Abdominal multi-organ segmentation with organ-attention networks and statistical fusion. Med. Image Anal. 55, 88–102 (2019).
[6] Noone, T. C., Semelka, R. C., Chaney, D. M. & Reinhold, C. Abdominal imaging studies: Comparison of diagnostic accuracies resulting from ultrasound, computed tomography, and magnetic resonance imaging in the same individual. Magn. Reson. Imaging 22, 19–24 (2004).
[7] Merkle, E. M. & Nelson, R. C. Dual gradient-echo in-phase and opposed-phase hepatic MR imaging: A useful tool for evaluating more than fatty infiltration or fatty sparing. Radiographics 26, 1409–1418 (2006).
[8] Shetty, A. S. et al. In-phase and opposed-phase imaging: Applications of chemical shift and magnetic susceptibility in the chest and abdomen. Radiographics 39, 115–135 (2019).
[9] Zhang, G. et al. Cross-modal prostate cancer segmentation via self-attention distillation. IEEE J. Biomed. Heal. Informatics 14, 1–1 (2021).
[10] Lin, M. et al. Fully automated segmentation of brain tumor from multiparametric MRI using 3D context deep supervised U-Net. Med. Phys. 48, 4365–4374 (2021).
[11] Dolz, J. et al. HyperDense-Net: A hyper-densely connected CNN for multi-modal image segmentation. IEEE Trans. Med. Imaging 38, 1116–1126 (2019).
[12] Zhang, Y. et al. Deep-learning based rectal tumor localization and segmentation on multi-parametric MRI. in ISMRM (2022).
[13] Zhou, C. et al. One-pass multi-task convolutional neural networks for efficient brain tumor segmentation. in MICCAI 11072, 637–645 (2018).
[14] Zhang, W. et al. Deep convolutional neural networks for multi-modality isointense infant brain image segmentation. Neuroimage 108, 214–224 (2015).
[15] Pinto, A. et al. Enhancing clinical MRI perfusion maps with data-driven maps of complementary nature for lesion outcome prediction. in MICCAI 11072, 107–115 (2018).
[16] Nie, D., Wang, L., Gao, Y. & Shen, D. Fully convolutional networks for multi-modality isointense infant brain image segmentation. in IEEE ISBI 1342–1345 (IEEE, 2016).
[17] Hazirbas, C. & Ma, L. FuseNet: Incorporating depth into semantic segmentation via fusion-based CNN architecture. in ACCV (2016).
[18] Li, C. et al. Adaptive convolution kernels for breast tumor segmentation in multi-parametric MR images. in ISMRM (2022).
Figures