3764
Automated Segmentation of Knee Cartilage from Ultra-High Resolution 7 Tesla 3D bSSFP MRI Using Transfer Learning1Imperial College London, London, United Kingdom
Synopsis
Keywords: Segmentation, Cartilage, Osteoarthritis
7T 3D knee MRI shows great promise for the quantification of cartilage volume and thickness to assess osteoarthritis, but manual segmentation is time consuming. Segmented 7T 3D knee datasets to train machine learning techniques are limited. Here, we trained a network on a larger dataset of segmented 3T 3D knee MRI scans and used transfer learning to create an automatic segmentation network of 7T MRI knee cartilage using a small number of segmented 7T images. The resulting network constructed demonstrated vastly improved automatic segmentation of the knee cartilage at 7T compared to the network trained with limited 7T data only.Introduction
7T MRI has been demonstrated to show great promise for musculoskeletal applications, and it was approved for clinical use for the first time in 2017[1]. Unlike 3T MRI, there are not yet large standardised datasets available at 7T. Thus, there are very few 7T MRI knee cartilage data available for convolutional neural network (CNN) training, leading to poor CNN performance. Transfer learning (TL) can effectively solve the problem that training data is expensive to acquire and, for the segmentation problem, very time consuming to produce[2]. We used TL to complete the training of an automatic segmentation network based on a large number of publicly available segmented knee cartilage data from the Osteoarthritis Initiative (OAI)[3]. The training network was also optimized by: (1) shuffling data, (2) removing MRI slices not containing knee cartilage, and (3) slicing 3D images along different sections to get 2D images for training.Once trained, quantitative metrics were used to evaluate the performance of the automatic segmentation network on the 7T MRI knee cartilage dataset.
Methods
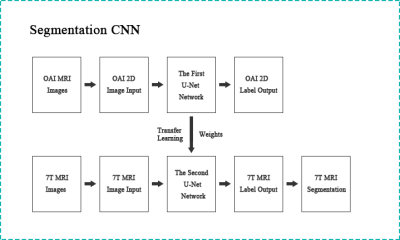
The architecture of the transfer learning system we used is shown in Figure 1. The transfer learning system can be divided into two parts, each of which consists of a two-dimensional CNN, which is the U-Net network model[4]. The training steps are as follows:- We first reformatted 3D images along the sagittal section into 2D images. The number of 2D sagittal section OAI slices was 28160 (protocol was 3D DESS). The number of slices for training set, validation set and test set was: 19200, 4480 and 4480. The number of 2D sagittal section 7T MRI slices was 1120 (protocol was phase-cycled bSSFP). All images were acquired with appropriate institutional informed consent. The number of slices for training set, validation set and test set was: 800, 160 and 160.
- The matrix size of OAI knee cartilage slices (384×384) is different from that of 7T MRI knee cartilage slices (512×512). We reconstructed the matrix size of OAI knee cartilage slices from (384×384) to (512×512) by nearest neighbour interpolation to obtain better automatic segmentation performance.
- The first U-Net model was then trained with reconstructed low-resolution OAI knee cartilage images, and the best trained U-Net weights were imported into the second U-Net model for use on 7T images.
- Based on these weights, the second U-Net model was trained with a small number of 7T MRI knee cartilage images to complete the fine-tuning of the model. After training, the second U-Net network is used for automatic segmentation of 7T MRI knee cartilage.
- Slice orders were shuffled for model training. This helps to avoid the model learning sequential features that are meaningless for knee cartilage segmentation during the training process. This optimisation method can reduce overfitting and increase the robustness of the model.
- Slices that did not contain cartilage were omitted from training. These slices do not provide meaningful features for knee cartilage segmentation, and will reduce accuracy of the model.
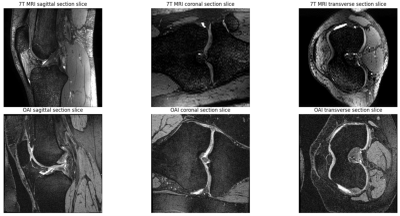
- 3D images were further reformatted along other two orientations, coronal and axial (transverse), and used as additional training data. Figure 2 shows the knee cartilage slices of different sections of OAI and 7T MRI.
Results
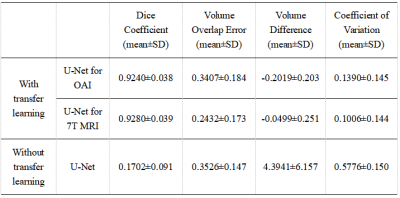
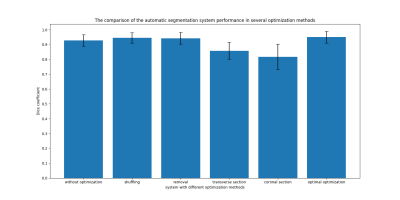
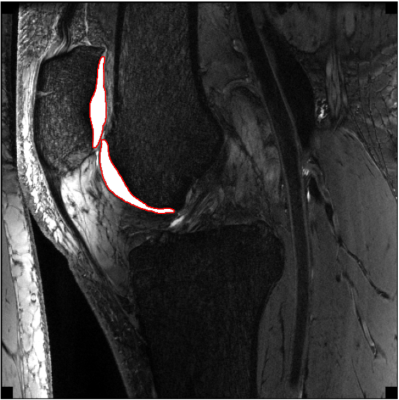
Figure 3 shows the results of DC, VOE, VD and CV for knee cartilage segmentation of the test set with and without transfer learning. It can be seen that the segmentation effect of the automatic segmentation network trained by the transfer learning system is much better than that of the automatic segmentation network trained directly by U-Net. In addition, Figure 4 shows a comparison of the automatic segmentation system performance in several optimisation methods we used. It can be seen that the effect of the automatic segmentation network is the best when the sagittal slices are used for training, the training slices are shuffled and the slices that do not contain knee cartilage are removed. Figure 5 shows an example of the trained transfer learning system segmenting a knee slice in sagittal section of a 39-year-old male subject.Disscusion and Conclusion
The results show that the 7T MRI knee cartilage automatic segmentation network trained using transfer learning performs much better than the automatic segmentation network trained without transfer learning. The final network at 7T outperforms segmentation accuracy of the original network trained on 3T data, probably due to the much higher quality of the 7T images. It also outperforms segmentation accuracy of other 3T-based networks in the literature, again likely due to the high resolution, SNR, and excellent contrast of the 7T images.Acknowledgements
First of all, I would like to extend my sincere gratitude to my supervisor, Neal Bangerter, for his instructive advice and useful suggestions on my project.
I am deeply grateful of his help in the completion of this project.I am also deeply indebted to all the other tutors and teachers in Department of Bioengineering for their direct and indirect help to me.
Special thanks should go to my friends who have put considerable time and effort into their comments on the draft.
Finally, I am indebted to my parents for their continuous support and encouragement.
References
1. Juras, Vladimir, et al. "Magnetic resonance imaging of the musculoskeletal system at 7T: morphological imaging and beyond." Topics in Magnetic Resonance Imaging 28.3 (2019): 125.
2. Weiss, Karl, Taghi M. Khoshgoftaar, and DingDing Wang. "A survey of transfer learning." Journal of Big data 3.1 (2016): 1-40.
3. WWW: Web page of Osteoarthritis Initiative Coordinating Centre. https://nda.nih.gov/oai/.
4. Ronneberger, Olaf, Philipp Fischer, and Thomas Brox. "U-net: Convolutional networks for biomedical image segmentation." International Conference on Medical image computing and computer-assisted intervention. Springer, Cham, 2015.
Figures