3761
Cardiac MR Image Segmentation in the Presence of Respiratory Motion Artifacts
1Biomedical Engineering Department, Eindhoven University of Technology, Eindhoven, Netherlands, 2MR R&D - Clinical Science, Philips Healthcare, Best, Netherlands
Synopsis
Keywords: Segmentation, Segmentation
Object motion during the acquisition of magnetic resonance images can negatively impact image quality by introducing inconsistencies in the k-space data and in turn, blurring and ghosting artifacts on the acquired images. Such artifacts represent significant challenges in the clinical deployment of automated segmentation algorithms. In this work, we present an ensemble of approaches aimed at developing a robust and generalizable segmentation model, particularly tailored to handle the appearance of respiratory motion artifacts. We achieve this by introducing k-space based simulation for augmentation, as well as by reducing common errors across basal and apical slices with a region-focused segmentation approach.
Introduction
Respiratory motion artifacts, commonly appearing during the acquisition of cardiac magnetic resonance (CMR) images, can negatively affect the quality of acquired images. In fact, respiratory motion introduces inconsistencies in k-space data between different segments, distributed over multiple heart beats, leading to fuzziness at the edges of the imaged tissue, as well as ghosting artifacts if the center of the k-space is affected [1]. If severe, such artifacts negatively impact the performance of automated segmentation models and restrict their deployment in clinical practice. In this work, we propose an ensemble of approaches aimed at developing a robust and generalizable segmentation method, particularly tailored to handle the appearance of respiratory motion artifacts in CMR images.Methods
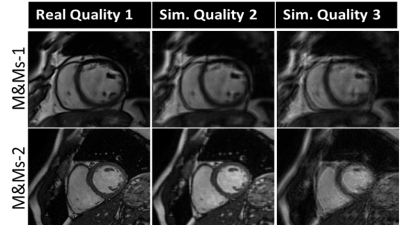
The proposed approach is developed on short-axis end-diastole (ED) and end-systole (ES) CMR images provided by the CMRxMotion challenge [2], consisting of 45 (25 for training and validation) healthy volunteers trained to simulate four levels of respiratory motion: (i) adhere to the breath-hold instructions, (ii) halve the breath-hold period, (iii) breathe freely and (iv) breathe intensively. All images of diagnostic quality are acquired from the same MRI scanner (Siemens 3T Magnetom Vida) and segmented into three label classes, i.e., the left ventricle (LV), left ventricle myocardium (MYO), and right ventricle (RV).The backbone of the proposed segmentation method is based on an ensemble of 2D nnU-Net [3] models, with substantial modifications for improving the model robustness to motion artifacts. To tackle the limitation in the number of training images, we utilize the open-source M&Ms-1 and M&Ms-2 data [4], augmented with their simulated, motion corrupted counterparts in addition to training images from the CMRxMotion challenge. Similar to prior work using k-space artifact simulation for brain and respiratory motion [5,6], we model breathing motion as a sinusoidal translation of the image in one direction, as a first approximation. We assume that the k-space data is acquired in multiple segments at different respiration points, corresponding to different amounts of translation. Thus, the combined k-space is composed of different sections from the k-space data for each translated image and transformed to the image domain via inverse Fourier transformation. The severity of the motion artifact is varied by controlling the period and the amplitude of the sinusoidal function, corresponding to the number of breathing cycles and the maximum organ translation during the acquisition time window, respectively.
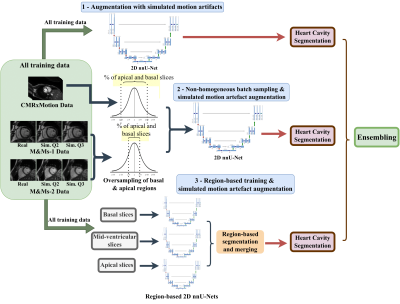
To address the difficulties in segmenting the basal and apical regions of the heart, particularly in the presence of motion artifacts, we utilize non-homogeneous batch sampling to over-sample basal and apical slices seen during training. Thus, for each mini-batch, slices from the apex and the base of CMRxMotion images are selected within a probability of 2σ of the mean in a normal Gaussian distribution, while the basal and apical slices of other datasets used for training are sampled within a probability of 1σ. We additionally employ a region-based training approach, by utilizing three separate nnU-Net models aimed at segmenting basal, mid-ventricular and apical slices, respectively. Both training and testing images are roughly split into different slices and merged back into an original 3D volume at inference time. In total, we obtain three sets of heart tissue segmentation maps, as seen in Figure 2, consisting of an ensemble of predicted softmax probabilities, acquired from (i) a model trained utilizing only augmentation with simulated motion artifacts; (ii) a model trained using simulated motion artifact augmentation and non-homogeneous batch sampling; and (iii) a combination of three region-focused segmentation models, all trained using simulated motion augmentation.
Results and Discussion
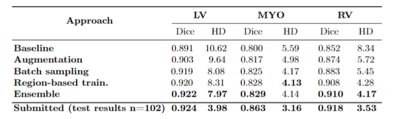
To assess the robustness of the proposed segmentation pipeline, we compare it against a baseline model, consisting of a regular nnU-Net framework trained with a combination of M&Ms-1, M&Ms-2 and CMRxMotion images. We additionally study the influence of different modules included in the final ensemble, as seen in Table 1. The obtained results suggest that augmentation with data containing simulated motion artifacts significantly improves the performance and adaptation of the segmentation model in the presence of motion artifacts, particularly across the myocardium and right-ventricle. However, focusing the training on the basal and apical regions of the heart by utilizing non-homogeneous batch sampling and region-based segmentation shows major improvements compared to other models. This confirms our assumption that slices around the apex and the base of the heart are the major source of segmentation errors, particularly due to stronger effects of motion artifacts in these regions causing blurring and ambiguous cavity borders. Finally, ensembling additionally reduces under-segmentation and improves prediction performance around region borders.Conclusion
This work demonstrates the effectiveness of k-space based motion simulation in handling data scarcity by simulating different levels of motion artifacts on artifact-free cardiac MR images. The addition of simulated motion corrupted images to the training helps the segmentation network adapt to artifacts appearing due to respiratory motion during acquisition by reducing typical segmentation errors caused by blurring, occluded and ambiguous tissue boundaries, and degraded image quality. In addition, we identify that motion artifacts affect the slices around the base and apex of the heart more severely, which we tackle by employing non-homogeneous sampling and region-based training, allowing us to better handle the variation in heart tissue shape and appearance.Acknowledgements
This research is a part of the OpenGTN project, supported by the European Union in the Marie Curie Innovative Training Networks (ITN) fellowship program under project No. 76446.
References
1] Ferreira, P.F., Gatehouse, P.D., Mohiaddin, R.H., Firmin, D.N.: Cardiovascular magnetic resonance artefacts. Journal of Cardiovascular Magnetic Resonance 15(1), 1–39 (2013)
[2] Wang, S., Qin, C., Wang, C., Wang, K., Wang, H., Chen, C., Ouyang, C., Kuang, X., Dai, C., Mo, Y. and Shi, Z., 2022. The Extreme Cardiac MRI Analysis Challenge under Respiratory Motion (CMRxMotion). arXiv preprint arXiv:2210.06385.
[3] Isensee, F., Jaeger, P.F., Kohl, S.A., Petersen, J., Maier-Hein, K.H.: nnu-net: a self-configuring method for deep learning-based biomedical image segmentation. Nature methods 18(2), 203–211 (2021)
[4] Campello, V.M., Gkontra, P., Izquierdo, C., Martin-Isla, C., Sojoudi, A., Full, P.M., Maier-Hein, K., Zhang, Y., He, Z., Ma, J., et al.: Multi-centre, multi-vendor and multi-disease cardiac segmentation: the m&ms challenge. IEEE Transactions on Medical Imaging 40(12), 3543–3554 (2021)
[5] Shaw, R., Sudre, C.H., Varsavsky, T., Ourselin, S., Cardoso, M.J.: A k-space model of movement artefacts: application to segmentation augmentation and artefact removal. IEEE transactions on medical imaging 39(9), 2881–2892 (2020)
[6] Oksuz, I., Ruijsink, B., Puyol-Antón, E., Clough, J.R., Cruz, G., Bustin, A., Prieto, C., Botnar, R., Rueckert, D., Schnabel, J.A., et al.: Automatic cnn-based detection of cardiac mr motion artefacts using k-space data augmentation and curriculum learning. Medical image analysis 55, 136–147 (2019)
[7] Abraham, N., Khan, N.M.: A novel focal tversky loss function with improved attention u-net for lesion segmentation. In: 2019 IEEE 16th international symposium on biomedical imaging (ISBI 2019). pp. 683–687. IEEE (2019)
Figures