3760
A deep neural network framework of few-shot learning with domain adaptation for automatic meniscus segmentation in 3D Fast Spin Echo MRI1CU Lab for AI in Radiology (CLAIR), Department of Imaging and Interventional Radiology, The Chinese University of Hong Kong, Hong Kong, China, 2Department of Radiology, Shanghai Sixth People's Hospital Affiliated to Shanghai Jiao Tong University School of Medicine, Shang hai, China, 3Department of Imaging and Interventional Radiology, The Chinese University of Hong Kong, Hong Kong, China, 4Department of Orthopaedics & Traumatology, The Chinese University of Hong Kong, Hong Kong, China, 5Philips Healthcare, Hong Kong, China, 6Guangdong Provincial People's Hospital, Guangdong Academy of Medical Sciences, Guang Zhou, China
Synopsis
Keywords: Segmentation, Machine Learning/Artificial Intelligence
Automatic meniscus segmentation is highly desirable for quantitative analysis of knee joint diseases. As three-dimensional Fast Spin Echo (3D FSE) is a promising MR (magnetic resonance) imaging technique to evaluate the tissues of the knee joint. In this study, we explore meniscal segmentation on 3D FSE MRI. Manually annotating 3D knee images is challenging since it is time-consuming and requires clinical expertise. In this study, we propose a domain adaption-based few-shot learning method for meniscal segmentation on 3D FSE images using only one annotated MRI data. We demonstrate that the proposed method outperformed the fully supervised segmentation model.Introduction
Accurate meniscus segmentation of MR images is important for quantitative morphology analysis of the meniscus and evaluation of knee joint pathology [1]. However, automatic fully-supervised segmentation models inevitably require a large number of annotated data. It is challenging to build a manually annotated meniscus dataset since it is time-consuming and requires clinical expertise. Recent developments of few-shot learning methods emphasize using only a few annotated data together with sufficient unannotated data for training. Three-dimensional (3D) Fast/Turbo Spin Echo(FSE/TSE) sequences are prevalent for visualizing knee structures because they can be acquired in 3D high isotropic resolution and reformatted into arbitrary planes with significantly reduced total scan time compared to 2D FSE [2]. In this work, we propose a novel few-shot learning framework with domain adaptation for automatic meniscus segmentation of 3D FSE with only one annotated data. Our experimental results show that our proposed framework outperforms state-of-the-art semi-supervised methods and the standard U-Net [3] in a fully supervised setting.Methods
The study was approved by the institutional review board. Datasets used in this study include A) 176 MR imaging examinations acquired using 3D double echo steady state (DESS) sequence with segmentation labels from the Osteoarthritis Initiative ( http://www.oai.ucsf. edu/ ) database; B) 30 proton density-weighted 3D FSE MR imaging scans acquired at our institution using VISTATM (Philips Healthcare, Best, Netherlands), with 24 used for training and 6 for testing. The imaging parameters of our 3D FSE were as follows: TR/TE 900/33.6 ms; 150 slices with an isotropic resolution of 0.8 × 0.8 × 0.8 mm; an echo train length of 42; and a SENSE acceleration factor of 2. The acquisition time is approximately 2.9 min. The reduced scan time meant that our acquisition data had a relatively low signal-to-noise ratio. Dataset B covers the full spectrum of Kellgren and Lawrence (KL) grades, including several cases with KL grade 4. Manual annotation of the meniscus from dataset B was carried out by two musculoskeletal radiologists using ITK-SNAP.Our proposed framework, depicted in Fig 1, is based on unsupervised domain adaptation (UDA) with mixup-augmented adversarial learning (MAL) and self-learning (SL). It consists of three segmentation networks (U1, U2, and U3) which are standard U-Net and two fully convolutional discriminators (D1 and D2). A MAL UDA method introduced in [4] is used to train U1 and D1 for adapting from DESS to FSE MR images. The annotated DESS MR images and unannotated FSE MR images are fed into the U1 to optimize U1 and D1. The idea of mixup is to linearly interpolate the inputs($$$x$$$) and the corresponding annotations ($$$y$$$) to construct paired virtual training data.[4] The virtual training data can be formulated as $$$x_{mix}=\lambda x+(1-\lambda)x_{perm}$$$ and $$$y_{mix}=\lambda y+(1-\lambda)y_{perm}$$$, in which $$$x_{perm},y_{perm}=permute(x,y)$$$. $$$\lambda\in [0,1]$$$ obeys a beta distribution $$$\lambda$$$ ~ $$$Beta(\alpha,\alpha)$$$, where $$$\alpha$$$ denotes the strength of augmentation and is set as 1. The segmentation loss with mixup can be defined as $$$L_{seg}=\lambda L_{dice}\left(p,y\right)+(1-\lambda)L_{dice}(p,y_{perm})$$$ , where $$$L_{dice}$$$ denotes dice coefficient loss and $$$p$$$ denotes predictions of the segmentation network. The weights of U1 are then shared with U2. Hung et al. [5] proposed an adversarial semi-supervised segmentation method with a specific semi-supervised loss. We introduced the mixup into this method to train U2 and D2. One annotated FSE MR imaging scan and 23 unannotated FSE MR imaging scans are used to optimize U2 and D2. Note D2 provides additional semi-supervised (few-shot) signals by confidence map which facilitates discovering the reliable regions in predictions of unannotated images. Lastly, we developed a teacher-to-student SL manner to further improve segmentation accuracy in U3. Both one annotated FSE MR imaging examinations and pseudo-labels from unannotated FSE generated by U2 are used to supervise U3.
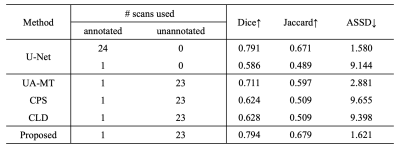
For comparison, we also trained a U-Net using supervised learning (24 annotated MR imaging examinations for training and 6 for testing), and several state-of-the-art semi-supervised methods including UA-MT [6], CPS [7], and CLD [8] utilizing the few-shot training strategy (1 annotated and 23 unannotated data for training).
Results & Discussion
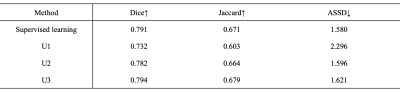
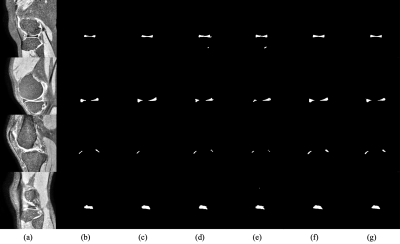
Table 2. Quantitative evaluations of the performances of U1, U2, U3, and the supervised learning. Note U1, U2, and U3 all achieve good performance for meniscal segmentation. The desired model U3 outperforms the supervised learning model. Table 3 shows the performance of our method compared to the other semi-supervised approach using few-shot learning. Example images are shown in Fig. 4. Note the proposed method achieves improved accuracy of meniscal segmentation compared to the other methods.Conclusions
We propose a novel few-shot learning framework with domain adaptation for meniscal segmentation of 3D FSE MR images. Our approach can be used to tackle challenges related to lack of annotated images for training. Our method can achieve satisfactory meniscal segmentation using 3D FSE with only one annotated dataset based on domain adaptation.Acknowledgements
This work is supported by a grant from the Innovation and Technology Commission of the Hong Kong SAR (Project MRP/001/18X).References
1. Rahman MM, Dürselen L, Seitz AM. Automatic segmentation of knee menisci - A systematic review. Artif Intell Med. 2020 May;105:101849. doi: 10.1016/j.artmed.2020.101849. Epub 2020 May 6. PMID: 32505421.
2. Gold GE, Busse RF, Beehler C, Han E, Brau AC, Beatty PJ, Beaulieu CF. Isotropic MRI of the knee with 3D fast spin-echo extended echo-train acquisition (XETA): initial experience. AJR Am J Roentgenol. 2007 May;188(5):1287-93. doi: 10.2214/AJR.06.1208. PMID: 17449772.
3. Ronneberger, Olaf, Philipp Fischer, and Thomas Brox. "U-net: Convolutional networks for biomedical image segmentation." International Conference on Medical image computing and computer-assisted intervention. Springer, Cham, 2015.
4. Panfilov, Egor, et al. "Improving robustness of deep learning based knee mri segmentation: Mixup and adversarial domain adaptation." Proceedings of the IEEE/CVF International Conference on Computer Vision Workshops. 2019.
5. Hung, Wei-Chih, et al. "Adversarial learning for semi-supervised semantic segmentation." arXiv preprint arXiv:1802.07934 (2018).
6. Yu, Lequan, et al. "Uncertainty-aware self-ensembling model for semi-supervised 3D left atrium segmentation." International Conference on Medical Image Computing and Computer-Assisted Intervention. Springer, Cham, 2019.
7. Chen, Xiaokang, et al. "Semi-supervised semantic segmentation with cross pseudo supervision." Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition. 2021.
8. Lin, Yiqun, et al. "Calibrating Label Distribution for Class-Imbalanced Barely-Supervised Knee Segmentation." arXiv preprint arXiv:2205.03644 (2022).
Figures