3746
Distinct contrast patterns in patient-derived tumor xenografts (PDX) mouse model revealed by pH-sensitive MR contrast agent
Akira Sumiyoshi1, Takahiro Nomoto2, Megumi Iiyama1, Kensuke Osada1, Rumiana Bakalova1, Nobuhiro Nishiyama2, and Ichio Aoki1
1National Institutes for Quantum Science and Technology, Chiba, Japan, 2Tokyo Institute of Technology, Yokohama, Japan
1National Institutes for Quantum Science and Technology, Chiba, Japan, 2Tokyo Institute of Technology, Yokohama, Japan
Synopsis
Keywords: Molecular Imaging, Cancer
Here we performed a molecular MR imaging study on patient-derived tumor xenografts (PDX) mouse model. In vivo molecular MR imaging demonstrated that proliferative vs. non-proliferative regions exist in PDX model and highly specific contrast agent such as PEGMnCap (pH-responsive nanoparticle based on Mn) can delineate active and hypoxic regions while small-molecular size contrast agent such as Gd-DOTA (gadoterate meglumine) can delineate non-active tumor regions. These MR image separations were not clearly obtained with a cancer cell line, suggesting more complex microenvironment of PDX model and may provide valid translational feedbacks for clinical practice.Introduction
There has been increasing interests in the development and characterization of patient-derived tumor xenograft (PDX) models for cancer research. PDX models mostly retain the basic histological and genetic characteristics of their donor cancer patients1. These models have been shown to predict clinical outcomes and are used for preclinical drug screening, biomarker identification, and personalized medicine strategies2. To our knowledge, no preclinical MR imaging study on PDX models has been reported in the literature, although these models may provide direct translational imaging evidence that may not be possible with cultured cells3. In this study, we obtained a commercially available PDX colon cancer mouse model and performed a molecular MR imaging study using MR contrast agents that have been developed in our institute for cancer research4,5.Methods
The PDX mouse model was purchased from the Central Institute for Experimental Animals, Kawasaki, Japan. The ID was COL-01-JCK that was established from male colon cancer patients without any drug treatment history. The original PDX mouse model was expanded to other athymic nude mice (BALB/c-nu/nu; SLC, Hamamatsu, Japan) using a tissue transplantation kit (Ez-Plant, Kyushu Organ Needle, Kumamoto, Japan) and a total of 10 PDX mouse models were used. MRI experiments were performed 4-8 weeks after transplantation once the tumor size was over 10-mm in diameter (Fig 1A). Mice were anesthetized with Isoflurane and MRI scans were acquired using either 7-tesla Biospec 70/40 or 70/20 with a cryogenically cooled RF coil (Bruker Biospin, Ettlingen, Germany). T1-weighted images were acquired with 3D-MPRAGE sequence using the following parameters: TR, 4000-ms; TE, 2.2-ms; inversion time, 1050-ms; resolution, 0.05-mm isotropic; NEX, 10; and scanning time, 30-minutes. T1-weighted scans were repeated before and after administration of contrast agents through the mouse tail vein. Gd-DOTA (gadoterate meglumine) and Gadolisome5 were purchased and PEGMnCap (Mn2+-doped calcium phosphate nanoparticles with a PEG shell) were prepared according to the previous paper4. Colon 26 mouse model was also prepared according to the previous paper for model comparison. Histopathological analysis such as hematoxylin and eosin (H&E) and Azan staining as well as immuno-stainings (Pimonidazole, Ki-67, and TUNEL) were performed from the same animals after MR imaging.Results
To characterize the tissue microenvironment of the PDX model, we adopted PEGMnCap that is pH-responsive MR contrast agent (Fig 1C). T1-weighted MRI before and after PEGMnCap administration (0.225 mmol/kg based on Mn) revealed that peripheral regions were selectively enhanced (Fig 1D) and histopathological analysis revealed that peripheral regions were indeed hypoxic and enriched with proliferation positive tumor cells (Fig 1E). TUNEL positive regions were not enhanced with PEGMnCap. To compare with conventional contrast agent, we adopted Gd-DOTA, where molecular size is around 1-nm, which were expected to leak from the vascular wall into the tumor. T1-weighted MRI before and after Gd-DOTA administration (0.4 mmol/kg based on Gd) revealed that central regions were selectively enhanced after 60-minutes of injection. Histopathological analysis revealed that central regions were enriched with TUNEL positive and without proliferation. Angiography imaging after Gadolisome administration (0.023 mmol/kg base on Gd) confirmed that there was no leakage from vessel wall (Fig 2C) as the average molecular size of Gadolisome was around 110-nm. When compared with classical Colon 26 model no such region-specific enhancement was observed (Fig 2D). High magnification image of histopathological analysis revealed that tumor vasculature was surrounded with collagen fiver, and TUNEL positive regions were found in the distal area from the vasculature (Fig 3A). Diffusion-based parametric image such as apparent diffusion coefficient (ADC) revealed that Gd-DOTA positive areas were not entirely overlapped with ADC image (Fig 3B), suggesting that Gd-DOTA positive regions were not fully accounted for water diffusion.Discussion
In vivo molecular MR imaging demonstrated that proliferative vs. non-proliferative regions exist in PDX model and highly specific contrast agent such as PEGMnCap can delineate active and hypoxic regions while small-molecular size contrast agent such as Gd-DOTA can delineate non-active tumor regions. These MR image separations were not clearly obtained with a cancer cell line, suggesting more complex microenvironment of PDX model and may provide valid translational feedbacks for clinical practice. Further preclinical MR studies such as extensions to other types of PDX model, adaptations of other types of contrast agent, and investigations on drug treatment response were expected.Acknowledgements
This work was supported by JSPS KAKENHI Grant Numbers JP21H04966, JP21H04966, JP21KK0201 and MRI devices were partly supported by JST/MEXT project for promoting public utilization of advanced research infrastructure, Grant Number JPMXS0450400422.References
- Tentler et al. (2012) Patient-derived tumour xenografts as models for oncology drug development. Nat Rev Clin Oncol 9:338–350.
- Gao et al. (2015) High-throughput screening using patient-derived tumor xenografts to predict clinical trial drug response. Nat Med 21:1318–1325.
- Ledford (2016) US cancer institute to overhaul tumour cell lines. Nature 530:391–391.
- Mi et al. (2016) A pH-activatable nanoparticle with signal-amplification capabilities for non-invasive imaging of tumour malignancy. Nat Nanotechnol 11:724–730.
- Nitta et al. (2018) Intratumoral evaluation of 3D microvasculature and nanoparticle distribution using a gadolinium-dendron modified nano-liposomal contrast agent with magnetic resonance micro-imaging. Nanomedicine 14:1315–1324.
Figures
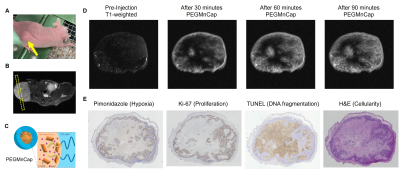
Fig. 1 (A) Athymic nude mouse with the tumor
xenograft (yellow arrow). (B) Slice position on the T2-weithed image. (C) Schematic
illustration of PEGMnCaP that consists of calcium phosphate core and PEG shell.
PEGMnCap releases Mn2+ through pH-responsive manner. (D) T1-weighted
images before and after PEGMnCap administration. (E) Histological evaluation of
the same subject. Pimonidazole, Ki-67, and TUNEL were stained in brown.
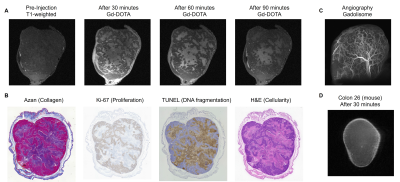
Fig. 2 (A) T1-weighted images before and after
Gd-DOTA administration. (B) Histological evaluation of the same subject. Collagen
fibers were stained in blue, nucleus and cytoplasm were stained in red using Azan
staining. Ki-67 and TUNEL were immuno-stained in brown. (C) MR angiography
using Gadolinium-contained liposome contrast agent (Gadolisome). (D) T1-weighted
images of mouse Colon 26 model after Gd-DOTA administration.
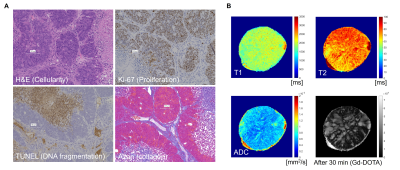
Fig. 3 (A) High magnification images of
histological staining. Tumor cells were partitioned by vasculature where
collagen fibers surround vessel wall. TUNEL positive cells were found in the distal
area from vasculature whereas Ki-67 positive cells were found in the proximal
area. (B) Multi-parametric and quantitative evaluation of PDX model. Gd-DOTA
positive area were not entirely overlapped with ADC-high areas.
DOI: https://doi.org/10.58530/2023/3746