3719
Rapid multi-slice whole-brain B1+-mapping at 7T using deep learning1Physikalisch-Technische Bundesanstalt, Berlin and Braunschweig, Germany, 2Division of Imaging Sciences and Biomedical Engineering, King's College London, London, United Kingdom, 3Technical University of Munich, Munich, Germany, 4Imperial College London, London, United Kingdom, 5Medical Physics in Radiology, German Cancer Research Center (DKFZ), Heidelberg, Germany, 6Center for Magnetic Resonance Research, University of Minnesota, Minneapolis, MN, United States
Synopsis
Keywords: RF Pulse Design & Fields, Parallel Transmit & Multiband
In this work we utilize deep learning to estimate multi-slice whole-brain B1+-maps in sub-seconds from initial localizer scans at 7T. The investigated neural networks use the receive profiles of the individual coil elements of an 8Tx/8Rx transceiver head coil as input information. The networks are trained on seven volunteers and tested in 2 unseen subjects for transversal/coronal/sagittal slices by comparing the prediction with the acquired B1+-maps. Subsequently, the feasibility of using the DL-based B1+-maps in a subject-specific calibration pipeline is demonstrated.Purpose
Subject-tailored parallel transmit (pTx) pulses for ultra-high fields (UHF) applications are typically calculated based on subject-specific data of all transmit (Tx) channels, which requires additional adjustment scans1. In the human brain, transmit magnetic field (B1+) maps based on pre-saturation acquisitions have been obtained in less than a minute with 8 Tx channels2. Yet, a higher resolution or an increase to 16 or 32 Tx channels3,4 will lead to calibration times of several minutes. Particularly for routine clinical scans, a fast, push-button, in-situ Tx field calibration embedded in the scanner's adjustment routine is highly desired. Deep learning (DL) based B1+-mapping techniques have shown promising results in addressing this need and reducing calibration times5,6.This study investigates the feasibility of utilizing a recently presented DL-based method applied to single slices in the heart6 for rapid, whole-brain relative B1+-mapping at 7T. The B1+-maps are derived in sub-seconds from an initial multi-slice localizer scan obtained in a CP+-like mode, typically acquired for planning at the beginning of each scan session. Hence, the neural networks (NNs) use the receive (Rx) profiles (B1-) of the individual coil elements of an 8Tx/8Rx transceiver head-coil as input information. The NNs are trained on seven volunteers and tested in two unseen subjects by comparing DL-predictions with acquired B1+-maps for different slice orientations.
Methods
Multi-slice B1+-maps (8-10 slices, depending on anatomy) were measured in 9 subjects on a 7T whole-body MRI system (Magnetom 7T, Siemens, Erlangen, Germany) with a custom-built 8Tx/8Rx head-coil array7 utilizing a hybrid approach8. First a small flip-angle 2D multi-slice GRE image (TR=100ms, TE=2.9ms, resolution=2x2mm2, FOV=256x192mm2, slice thickness=4mm, nominal FA=10°, 15 slices) is obtained with all Tx channels transmitting using a default shim (i.e., the localizer). Then, the acquisition is repeated 8 times where only a single Tx channel is active each. The latter scans are then merged with a 3D actual flip-angle image (TR=75ms, TE=1.9ms, resolution=2x2x4mm3, FOV=256x192mm2, slice thickness=4mm, nominal FA=60°) for all Tx coils active resulting in the ground truth (GT) multi-slice B1+-maps.The Rx-channel-wise localizer is split into real and imaginary parts and stacked with the magnitude image along the channel dimension, forming the NN's input resulting in an input size of 128x96x17 per slice. Similarly, the GT Tx-channel-wise B1+-maps are split up into real and imaginary data and arranged to a size of 128x96x16. Normalizing the input and output gives the originally absolute data a relative nature. However, biases of relative methods (e.g., T1/proton density)9 are avoided by this procedure.
The investigated NNs are modified UNets10,4 with a symmetric loss11 function. The first NN (NNtra) was trained on 66 transversal slices from seven volunteers and tested on 17 transversal slices from two unseen subjects. The second (NNcor) was trained on 86 coronal and tested on 17 coronal slices, and the third (NNall) was trained on 219 transversal/coronal/sagittal slices and tested on 51 transversal/sagittal/coronal slices. The training parameters are as follows: ADAM optimizer, learning rate=1*10-4, 2000 epochs, batch size=2. DL is performed on a 24 GB NVIDIA Titan RTX. The predicted (PR) maps are qualitatively compared to the GT evaluated by the root mean squared error (RMSE) and structural similarity index measure (SSIM). Static (phase-only) and dynamic (4kT points) pTx optimization was subsequently performed on the DL-based B1+-maps for different optimization targets.
Results and Discussion
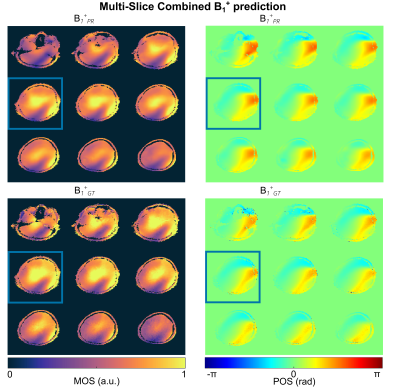
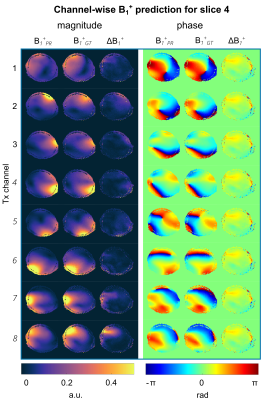
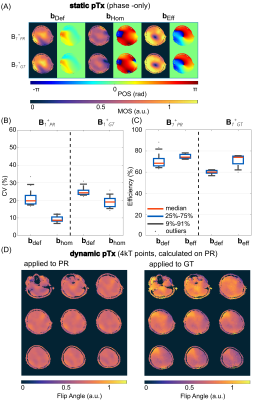
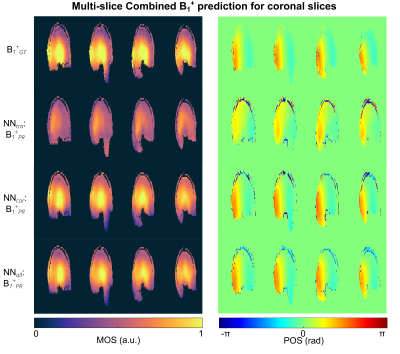
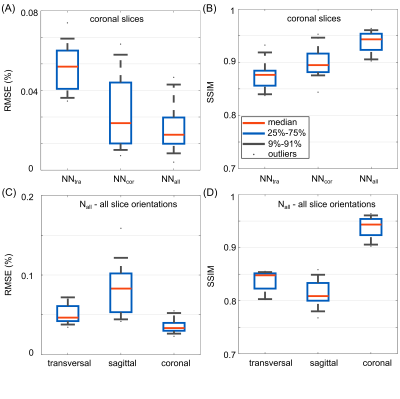
Fig.1 shows the magnitude and phase of the combined, multi-slice B1+-maps for the GT and PR by NNtra trained on multi-slice data in transversal orientation for one unseen test subject. The maps reflect the default shim setting bdef (no additional RF phases) resembling a CP+-mode. The DL-based B1+-maps follow the GT for magnitude and phase (RMSE=0.053±0.010%). The match between PR compared to the GT B1+-maps is further highlighted when looking at the channel-wise data for a central slice (Fig.2). Fig.3 depicts the B1+-shimming results for the same example slice after calculating a homogeneous shim vector bhom and an efficiency setting beff. The latter is highly useful, e.g., for subcutaneous fat saturation. The optimizations are based on the PR B1+-maps and the resulting setting is applied to the PR and GT B1+-maps. The GT and PR maps always reflect all features in the shimmed cases and for the default shim bdef . The feasibility of using DL-based B1+-maps in a subject-specific calibration pipeline is demonstrated by a quantitative evaluation for the shim vectors bhom and beff (Fig.3 (B)/(C)). This approach even provides good results for whole-brain dynamic pTx (4kt-points) excitation (Fig.3 (D)). Fig.4 shows the combined, multi-slice B1+-maps for four coronal slices for the GT and predicted by NNtra, NNcor, and NNall. While NNtra trained on transversal slices fails to predict coronal slices accurately, NNcor and NNall successfully predict the combined maps. An evaluation of the PR and the GT combined B1+-maps is shown for coronal slices for all NNs (Fig.5 (A)/(B)) and for all slice orientations predicted by NNall (Fig.5 (C)/(D)).Conclusion
Our results indicate the feasibility of estimating complex, channel-wise, multi-slice B1+-maps in the human head at 7T from a quick GRE localizer scan in multiple orientations. Although we relied on relative B1+-maps due to the normalization of the training data, an expansion to absolute data may be feasible.Acknowledgements
We gratefully acknowledge funding from the German Research Foundation (GRK2260-BIOQIC).References
1) Padormo F, Beqiri A, Hajnal JV, Malik SJ. Parallel transmission for ultrahigh-field imaging. NMR Biomed. 2016;29:1145-1161.
2) Herrler J, Liebig P, Gumbrecht R, Ritter D, Schmitter S, Maier A, Schmidt M, Uder M, Doerfler A, Nagel AM. Fast online-customized (FOCUS) parallel transmission pulses: A combination of universal pulses and individual optimization. Magn Reson Med. 2021; 85:3140–3153.
3) Orzada S, Solbach K, Gratz M, Brunheim S, Fiedler TM, Johst S, et al. A 32-channel parallel transmit system add-on for 7T MRI. Plos one. 2019; 14(9):e0222452.
4) Schmitter S, Wu X, Auerbach EJ, Adriany G, Pfeuffer J, Hamm M, Ugurbil K, Van de Moortele PF. Seven-tesla time-of-flight angiography using a 16-channel parallel transmit system with power-constrained 3-dimensional spoke radiofrequency pulse design. Invest Radiol. 2014; 49(5):314–325.
5) Eberhardt B, Poser BA, Shah NJ, Felder J. B1 field map synthesis with generative deep learning used in the design of parallel-transmit RF pulses for ultra-high field MRI. Z. Med. Phys. 2022; 1–12.
6) Krueger F, Aigner C, Hammernik K, Dietrich S, Lutz M, Menger J, Schaeffter T, Schmitter S. Rapid estimation of 2D relative B1+-maps from localizers in the human heart at 7T using deep learning. Magn Reson Med. 2022; 1-14.
7) Seifert F, Pfeiffer H, Mekle R, Waxmann P, Ittermann B. 7T 8-channel pTx head coil with high B1+ efficiency optimized for MRS. In: Proceedings of the 24th Annu. Meet. ISMRM, Singapore, Singapore. Abstract 3545, 2016.
8) Van de Moortele PF, Snyder C, DelaBarre L, Adriany G, Vaughan T, Ugurbil K. Calibration Tools for RF Shim at Very High Field with Multiple Element RF Coils: from Ultra Fast Local Relative Phase to Absolute Magnitude B1+ Mapping. In: Proceedings of the 15th Annual Meeting of ISMRM, Berlin, Germany: Abstract 1676; 2007.
9) Dietrich S, Aigner CS, Kolbitsch C, Mayer J, Ludwig J, Schmidt S, Schaeffter T, Schmitter S. 3D Free-breathing multichannel absolute B1+ Mapping in the human body at 7T. Magn Reson Med. 2021;85:2552–2567.
10) Ronneberger O, Fischer P, Brox T. U-Net: Convolutional Networks for Biomedical Image Segmentation. MICCAI. 2015;9351:234-241.
11) Terpstra ML, Maspero M, Sbrizzi A, van den Berg CAT. ⊥-Loss: a Symmetric Loss Function for Magnetic Resonance Imaging Reconstruction and Image Registration With Deep Learning. Med. Image Anal. 2022;80:102509.
Figures