3713
Impact of sampling strategies and residual U-net reconstruction on preserving high spatial frequencies in accelerated low-field MRI1Center for Adaptable MRI Technology (AMT center), Department of Biomedical Engineering, University of Basel, Allschwil, Switzerland, 2AMT center, Institute of Medical Sciences, School of Medicine, Medical Sciences & Nutrition, University of Aberdeen, Aberdeen, United Kingdom
Synopsis
Keywords: Machine Learning/Artificial Intelligence, Image Reconstruction, undersampling, averaging, data sampling
Low signal-to-noise (SNR) ratios inherent to low-field (LF) MRI challenge its relevance in clinical applications. Accelerating the acquisition by undersampling k-space followed by reconstruction techniques has already shown promising results. Yet, undersampling is usually done by skipping high-frequency information which can lead to misdiagnosis as small lesions can be missed. In this study, we exploited a specificity of low-SNR regimes, that is signal averaging, to explore different acceleration strategies without skipping crucial information in k-space. The DL-reconstructed images arising from those sampling schemes have been evaluated on acquired in-vivo and ex-vivo LF-MRI datasets, showcasing high-frequency preservation and potential for generalization.Introduction
Largely impeded by inherent lower spin polarization, an on-going challenge in low-field (LF) MRI research is to harvest sufficient Signal-to-Noise-Ratio (SNR) without compromising on scan duration. Efficient k-space sampling strategies are prime to reduce acquisition times, and undersampling (US) is thus often leveraged. The latter is done according to a sampling scheme (mask) that usually favours low frequencies defining the contrast and the overall object shapes in an image, at the expense of high frequencies containing small features (i.e., details). Undersampling is generally followed by a reconstruction method, such as deep learning (DL)1,2,3,4, to correct for the induced artefact. Nevertheless, it was shown that even the best performing models can miss small features that could be particularly relevant in clinical diagnostic settings5. In this study, we exploited a specificity of low-SNR regimes, that is signal averaging, to explore different sampling approaches for accelerated LF MR acquisitions. DL reconstruction was evaluated for three different down-sampling schemes providing 4-fold acceleration in datasets acquired at 0.1 T (4.2 MHz).Materials and Methods
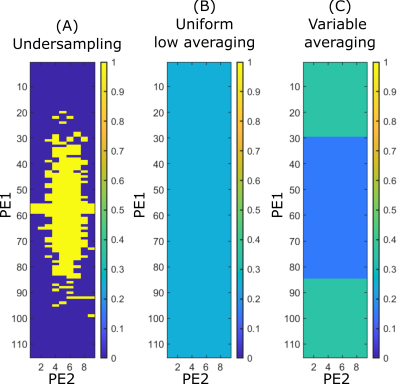
Sampling masks: Maintaining constant acquisition time, 25% sampling of a full 3D k-space with a Gaussian probability density function was challenged with uniform and variable averaging schemes of fully sampled k-spaces (cf. figure 1). Details on the investigated sampling masks are given below:1- Undersampling (US): it is a binary mask applied to phase encode 1 and 2 (PE1, PE2) tables (readout always fully sampled) following a Gaussian-like sampling pattern. 25% of the k-space is sampled and each sampled k-space line is acquired with maximum number of averages Nmax.
2- Uniform low averaging: every k-space line is averaged equally N times = 0.25xNmax.
3- Variable averaging (VA): Different number of averages are assigned to PE1 and PE2 steps in k-space6,7. Low frequencies are less averaged (0.15xNmax), as opposed to high frequencies (0.35xNmax), considering the signal associated with low frequencies is inherently higher. The rationale behind this approach is first to preserve high spatial frequencies, and redirect the DL problem from recovering missed information to denoising and recovering contrast.
Training: A total of 14 datasets of 3D MR in-vivo human wrist were acquired at 0.1 T (4.25 MHz) in a biplanar MRI system8 using a single transceiver coil. The acquisition matrix was set to 128×115×9. Imaging included mostly gradient echo (GRE) sequences with a couple of balanced steady state free precession (bSSFP) scans, with heterogenous acquisition parameters but a comparable overall SNR of 38±8. Four-fold accelerated acquisitions were simulated. For US, the fully acquired k-space was multiplied by a binary US mask. For uniform and variable averaging schemes, each k-space line was processed according to the assigned number of averages: the signal intensity was rescaled, and synthetic Gaussian noise was then added separately to the real and imaginary parts. The full and down-sampled k-spaces were normalized and inverse-Fourier transformed. Ultimately, three residual U-net9 models were trained on pairs of full and down-sampled MR images using the RMSProp optimizer with the mean squared error as a loss function. Data augmentation was applied to prevent overfitting.
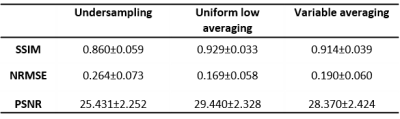
Testing: Performance of the DL models for the different masks were evaluated on four in-vivo human hand/wrist and three ex-vivo lamb lungs datasets. Full k-spaces were acquired with identical matrix dimensions as described above, with both GRE and bSSFP sequences, heterogeneous imaging parameters, and different Nmax, yet comparable overall SNR of 47±7. Each average was individually stored in a fourth dimension, allowing retrospective manipulation of k-spaces to generate images according to different masks. The reconstructed images were evaluated using the SSIM, normalized root MSE (NRMSE) and PSNR as metrics.
Results
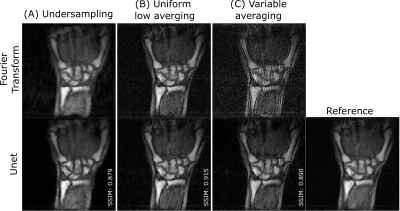
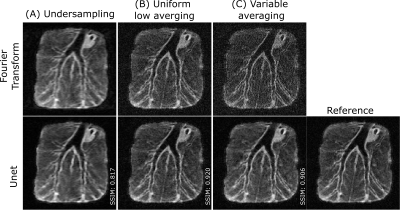
Figure 2 and 3 compare Fourier transform and residual U-net reconstruction in selected image samples in the human wrist and ex-vivo lamb lungs. Uniform and VA show good fidelity to the reference image. Despite an improvement in edge sharpness with DL reconstruction, US sampling inherently exhibits filtered high frequencies (i.e., blurring). Quantitatively, uniform averaging shows overall the best metrics (cf. table 1).Discussion
The three models were able to reconstruct ex-vivo lung images despite being trained on relatively small sets of wrist data, demonstrating good generalization of our training method. With residual U-net reconstruction, uniform or VA sampling seems more beneficial than undersampling skipping high frequency lines in k-space. The anticipated higher performance of VA sampling is not obvious when compared to uniform sampling. We hypothesize that this may result from a general lack of SNR where +10% averaging might not be sufficient for a low-intensity high-frequency signals to emerge from the noise level. Besides, it is worth noting that a model trained on uniform averaging is more generalizable than VA or US, adding a varying parameter inherent to the mask.Conclusion
In this work, we investigated three different down-sampling approaches followed by a residual U-net DL reconstruction. The results show that uniform and variable sampling are more beneficial than undersampling. The second underlying conclusion is also that residual U-net model probably performs best as a denoiser rather than in the retrieval of lost high-frequency features.Acknowledgements
Forschungfunds Grant from University of Basel (HIFI project).
Swiss National Science Foundation Grant No. 186861.
Swiss National Science Foundation Grant No. 198905.
References
1Hyun et al., Physics in Medicine and Biology, 63.13 (2018). https://doi.org/10.1088/1361-6560/aac71a
2Hammernik et al., Magnetic Resonance in Medicine, 79.6 (2018). https://doi.org/10.1002/mrm.26977
3Zhu et al., Nature, 555.7697
(2018). https://doi.org/10.1038/nature25988
4Ayde et al., Scientific Reports, 12.11394 (2022). https://doi.org/10.1038/s41598-022-14039-7
5Cheng et al., Proc of Machine Learning Research 121:121-135 (2020).
6Schoormans et al., Phys Med Biol, 65(4):045004 (2020). https://doi.org/10.1088/1361-6560/ab63b7
7Fiorito et al., NMR in Biomedicine, (2022). https://doi.org/10.1002/nbm.4826
8Constantinesco et al., Magnetic Resonance in Chemistry, 35 (1997), 69–75. https://doi.org/10.1002/(sici)1097-458x(199712)35:13<s69::aid-omr198>3.3.co;2-x.
9Ronneberger et al.,Medical Image Computing and Computer-Assisted Intervention – MICCAI 2015, (2015).
Figures