3710
A Faithful Deep Sensitivity Estimation Makes High-quality MRI Reconstruction1Department of Electronic Science, Biomedical Intelligent Cloud R&D Center, Fujian Provincial Key Laboratory of Plasma and Magnetic Resonance, National Institute for Data Science in Health and Medicine, Xiamen University, Xiamen, China, 2Department of Radiology, Zhongshan Hospital (Xiamen), Fudan University, Xiamen, China, 3Department of Radiology, Xiamen University, Xiamen, China, 4Department of Radiology, Zhongshan Hospital affiliated to Xiamen University, Xiamen, China, 5School of Computer and Information Engineering, Xiamen University of Technology, Xiamen, China
Synopsis
Keywords: Machine Learning/Artificial Intelligence, Machine Learning/Artificial Intelligence
Recent deep learning is superior in providing high-quality images and ultra-fast reconstructions in accelerated magnetic resonance imaging (MRI). Faithful coil sensitivity estimation is vital for MRI reconstruction. In this work, we propose a Joint Deep Sensitivity estimation and Image reconstruction network (JDSI). During the image artifacts removal, it gradually provides more faithful sensitivity maps, leading to greatly improved image reconstructions. Results on in vivo datasets and radiologist reader study demonstrate that, the proposed JDSI achieves the state-of-the-art performance visually and quantitatively, especially when the accelerated factor is high. Besides, JDSI also owns nice robustness to abnormal subjects.Purpose
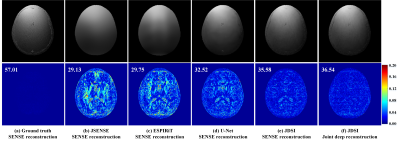
MRI is a leading diagnostic modality in modern medical science [1]. However, MRI scans usually require long time cost, and thus fast MRI technology attracts extensive research interests [2]. Although deep learning has demonstrated their superior performance in fast MRI on excellent image quality and reconstruction speed [3-7], most of them still rely on pre-estimated sensitivity maps and ignore their inaccuracy, resulting in the significant quality degradation of reconstructed images. Here, we clearly show that a more faithful sensitivity map leads to a lower reconstruction error, no matter for the conventional or deep learning methods (Figure 1). Thus, a joint deep network, called JDSI, is proposed to simultaneously achieve the faithful estimation of sensitivity maps and reconstruction of high-quality images.Method
The network architecture of our proposed JDSI is inspired by the alternating iterative idea of the joint conventional reconstruction [8], and JDSI continuously updates the sensitivity maps $$$\bf S$$$ with the vectorized reconstructed image $$$\bf x$$$ . Figure 2 gives the network architecture of our proposed JDSI. Specifically, our network firstly estimates initialized sensitivity maps from the undersampled multi-coil k-space and provides the initialized coil-combined image. Second, the sensitivity estimation module is utilized to refine sensitivity maps from the whole multi-coil k-space. Third, the undersampling artifacts are removed through the image reconstruction module. Fourth, reconstructed images and sensitivity maps are projected back to the k-space and then forced to maintain the data consistency to the acquired k-space. After repeating the last three modules $$$K$$$ times, the final reconstructed image can be obtained.To understand the behavior of the network, the mutual promotion of sensitivity estimation and image reconstruction is revealed through the visualization of network intermediate results (Figure 2(a)). The sensitivity maps gradually approximate the ground truth, while image artifacts are gradually removed. This enables our network to obtain faithful sensitivity maps and benefits high-quality image reconstruction.
In our implementation, the overall number of network phases is set as 5. The network is trained by minimizing the loss function which contains the mean square error from multi-coil images, combined-coil images, and sensitivity maps.
Results
Two brain datasets from the publicly shared fastMRI database [9] are used in our experiments. The proposed JDSI is compared with the state-of-the-art conventional method pFISTA-SENSE [10], and three deep learning methods including MoDL [11], Joint-ICNet [12], and HDSLR [13].To quantitatively evaluate the reconstruction performance, two objective criteria including peak signal-to-noise ratio (PSNR) and structural similarity index (SSIM) [14] are utilized. The higher PSNR and SSIM indicate less image distortions and better detail preservation in reconstructions, respectively.
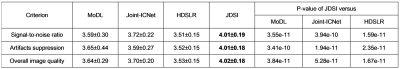
Besides, we further invite three radiologists (with 12, 29, and 30 years of clinical experience) to independently evaluate the reconstruction images from a diagnostic perspective. Three clinical concerned criteria including signal-to-noise ratio, artifacts suppression, and overall image quality are used. The score of each criterion has a range from 0 to 5 with precision of 0.1 (i.e., 0~1: Non-diagnostic; 1~2: Poor; 2~3: Adequate; 3~4: Good; 4~5: Excellent). The reader study is conducted online through our Cloud Brain Imaging platform [7, 15] at https://csrc.xmu.edu.cn/CloudBrain.html.
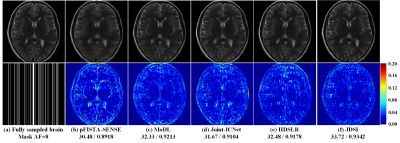
(i) Reconstruction of high acceleration factor (AF): Figure 3 shows that, for 1D Cartesian undersampling with AF=8, all compared methods yield results exhibiting obvious artifacts, whereas JDSI has smallest reconstruction error and outperforms them about 1.24~3.24dB in PSNR. These results demonstrate that, JDSI outperforms other compared methods both visually and quantitatively, indicating the excellent ability of artifacts suppression and details preservation.
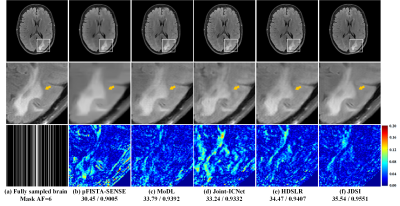
(ii) Reconstruction of abnormal subjects: Exploring the robustness of the proposed method to abnormal subjects is essential for clinical applications. Here, we use a brain dataset of healthy subjects to train all deep learning methods, then to reconstruct a brain image with white matter lesion. Figures 4(a) and (f) show that JDSI recovers the abnormal tissues (Marked with yellow arrows) most like the fully sampled image. Whereas other compared methods not recover the lesion well, resulting in over-smooth, structure loss, intensity loss, and residual artifacts (Figures 4(b)-(e)). It demonstrates that JDSI owns robustness to abnormal subjects.
(iii) Reader study: Figure 5 shows that, JDSI obtains highest mean scores. It is the only one which all three criteria are slightly over 4, indicating the reconstructed images are suitable for diagnosis. Besides, the differences between JDSI and other compared methods are great significant according to all P-values of the Wilcoxon signed rank test < 0.01. Thus, the image quality improvements obtained by JDSI is significant and the overall quality steps into the excellent level for clinical diagnosis.
Conclusion
In this work, we present a Joint Deep Sensitivity estimation and Image reconstruction network (JDSI) for accelerated magnetic resonance imaging (MRI). It simultaneously achieves the faithful estimation of sensitivity maps and the reconstruction of high-quality images. Results on in vivo datasets and radiologist reader study demonstrate that, JDSI provides improved and more robust reconstruction performance than state-of-the-art methods, especially when the acquisition is highly accelerated.Acknowledgements
See more details in the full-length preprint: https://arxiv.org/abs/2210.12723. This work was supported in part by the National Natural Science Foundation of China under grants 62122064, 61971361, 61871341, and 62071405, the Natural Science Foundation of Fujian Province of China under grant 2021J011184, the President Fund of Xiamen University under grant 0621ZK1035, and the Xiamen University Nanqiang Outstanding Talents Program. The authors thank Yirong Zhou and Jiayu Li for supporting the reader study on the Cloud Brain Imaging platform; Xinlin Zhang for the helpful discussions; Weiping He, Shaorong Fang, and Tianfu Wu from Information and Network Center of Xiamen University for the help with the GPU computing; Drs. Michael Lustig, Mathews Jacob, and Justin P. Haldar for sharing their codes online.
The correspondence should be sent to Prof. Xiaobo Qu (Email: quxiaobo@xmu.edu.cn)
References
[1] G. Harisinghani Mukesh, A. O’Shea, and R. Weissleder, "Advances in clinical MRI technology," Science Translational Medicine, vol. 11, no. 523, p. eaba2591, 2019.
[2] M. Lustig, D. Donoho, and J. M. Pauly, "Sparse MRI: The application of compressed sensing for rapid MR imaging," Magnetic Resonance in Medicine, vol. 58, no. 6, pp. 1182-1195, 2007.
[3] S. Wang et al., "Accelerating magnetic resonance imaging via deep learning," in IEEE 13th International Symposium on Biomedical Imaging (ISBI), 2016, pp. 514-517.
[4] B. Zhu, J. Z. Liu, S. F. Cauley, B. R. Rosen, and M. S. Rosen, "Image reconstruction by domain-transform manifold learning," Nature, vol. 555, no. 7697, pp. 487-492, 2018.
[5] T. Lu et al., "pFISTA-SENSE-ResNet for parallel MRI reconstruction," Journal of Magnetic Resonance, vol. 318, p. 106790, 2020.
[6] Z. Wang et al., "One-dimensional deep low-rank and sparse network for accelerated MRI," IEEE Transactions on Medical Imaging, DOI: 10.1109/TMI.2022.3203312, 2022.
[7] Q. Yang, Z. Wang, K. Guo, C. Cai, and X. Qu, "Physics-driven synthetic data learning for biomedical magnetic resonance: The imaging physics-based data synthesis paradigm for artificial intelligence," IEEE Signal Processing Magazine, DOI: 10.1109/MSP.2022.3183809, 2022.
[8] L. Ying and J. Sheng, "Joint image reconstruction and sensitivity estimation in SENSE (JSENSE)," Magnetic Resonance in Medicine, vol. 57, no. 6, pp. 1196-1202, 2007.
[9] F. K. Zbontar et al., "FastMRI: An open dataset and benchmarks for accelerated MRI," arXiv: 1811.08839, 2019.
[10] X. Zhang et al., "A guaranteed convergence analysis for the projected fast iterative soft-thresholding algorithm in parallel MRI," Medical Image Analysis, vol. 69, 101987, 2021.
[11] H. K. Aggarwal, M. P. Mani, and M. Jacob, "MoDL: Model-based deep learning architecture for inverse problems," IEEE Transactions on Medical Imaging, vol. 38, no. 2, pp. 394-405, 2019.
[12] Y. Jun, H. Shin, T. Eo, and D. Hwang, "Joint deep model-based MR image and coil sensitivity reconstruction network (Joint-ICNet) for fast MRI," in IEEE Conference on Computer Vision and Pattern Recognition (CVPR), 2021, pp. 5270-5279.
[13] A. Pramanik, H. Aggarwal, and M. Jacob, "Deep generalization of structured low-rank algorithms (Deep-SLR)," IEEE Transactions on Medical Imaging, vol. 39, no. 12, pp. 4186-4197, 2020.
[14] W. Zhou, A. C. Bovik, H. R. Sheikh, and E. P. Simoncelli, "Image quality assessment: from error visibility to structural similarity," IEEE Transactions on Image Processing, vol. 13, no. 4, pp. 600-612, 2004.
[15] Y. Zhou et al., "XCloud-pFISTA: A medical intelligence cloud for accelerated MRI," in 43rd Annual International Conference of the IEEE Engineering in Medicine & Biology Society (EMBC), 2021, pp. 3289-3292.
[16] M. Uecker et al., "ESPIRiT—An eigenvalue approach to autocalibrating parallel MRI: Where SENSE meets GRAPPA," Magnetic Resonance in Medicine, vol. 71, no. 3, pp. 990-1001, 2014.
[17] A. Sriram et al., "End-to-End variational networks for accelerated MRI reconstruction," in Medical Image Computing and Computer Assisted Intervention (MICCAI), 2020, pp. 64-73.
Figures