3702
Predicting Abdominal MRI Protocols using Electronic Health Records
Peyman Shokrollahi1, Juan M. Zambrano Chaves1, Avishkar Sharma1, Jonathan P.H. Lam1, Debashish Pal2, Naeim Bahrami2, Akshay S. Chaudhari1, and Andreas M. Loening1
1Radiology, Stanford University, Stanford, CA, United States, 2GE Healthcare, Sunnyvale, CA, United States
1Radiology, Stanford University, Stanford, CA, United States, 2GE Healthcare, Sunnyvale, CA, United States
Synopsis
Keywords: Machine Learning/Artificial Intelligence, Modelling, MR Radiology Workflow
Inaccurate selection of MRI protocols can impede diagnostics and therapeutic workflows, delay appropriate treatment, increase misdiagnosis likelihoods, and increase healthcare costs. Here we depict a machine-learning (ML) based system to accurately predict abdominal MR protocols, trained on the electronic medical records from 11,251 MR exam orders on 6,882 patients. The best model achieved a cumulative F1-score of 95.6% for top-three most-often-ordered protocols and a top-one F1 score of 78.5%. The proposed system can guide radiologists to appropriate protocol selections quickly, optimize workflows, and improve diagnostic accuracy, thereby by serving to support optimal patient outcomes.Introduction
More than 40 million MRI scans are conducted in the USA each year1. To carry out physicians’ orders, radiology personnel are tasked with selecting a protocol, inclusive of selecting an anatomical region (e.g., abdomen), focus (e.g., liver), and potential adjunct intervention (e.g., intravenous contrast). Current manual MRI protocol selection is time-consuming, cumbersome, and carries a risk of human error2. While machine learning (ML) applications have produced MRI acquisition and interpretation algorithms, relatively little such work has been applied to pre-image acquisition tasks, such as protocol selection2,3. Prior work has focused on a subset of MRI protocols (neuroradiology), or on the diagnosis/reason for exam only, leading to limited performance and usability. ML enhancement and optimization of the radiologist workflow can facilitate appropriate protocol selection, thereby improving patient safety and preventing inappropriate selections, which would otherwise risk incomplete diagnostic processes that can impede diagnostics and therapeutic workflows, delay appropriate treatment, increase misdiagnosis likelihoods, and increase healthcare costs and times2,4. This system differs from prior attempts to address this problem in that it does not rely on text inputs5,6. Here, we report the development and testing of an electronic medical record (EMR) database-trained ML technique that predicts rank-ordered abdominal MR protocols.Methods
We obtained EMRs for patients who underwent abdominal or pelvic MRI scans between 2017 and 2019 at a tertiary medical care center. The input signal consisted of a tabular EMR dataset included protocol-relevant data (e.g., radiology title, order priority, previous physician orders, order changes, procedure description, and diagnosis type), demographics (e.g., race, sex, ethnicity, height, weight, smoking status, insurance provider), admission information (e.g., age, obstetric status, patient level care, and patient service), prior laboratory (e.g., eGFR and urine creatinine), and patient history (e.g., allergies and medication history) . Laboratory values were lemmatized by grouping them into functionally redundant clinical measures (e.g., ‘urine creatinine’ and ‘creatinine, urine’). We chose the five most commonly ordered protocols for this analysis (Fig. 1b, Gadavist, Eovist, Prostate Local, MRCP, and Renal Adrenal). Extracted data were derived from 19,590 patients (some with multiple records), consisting of 38,010 exam order records with 48 EMR-based attributes. The data were preprocessed by filtering for age (<18 years or >90 years excluded), duplicate records, case non-relation to any of the top five radiology protocols, attributes with more than 70% missing values, and features that would not be available at inference time. The input signal was formed with records related to 11,251 exam orders from 6,882 separate patients and 37 attributes. The data were then shuffled and split into training (85%) and test (15%) by patient. The training set was also shuffled and split into training (85%) and validation (15%) by patient.Three ML decision-tree models7 informed by the aforementioned features were used to predict MRI protocols among the 5 protocols included: light gradient boosting machine (LGBM), extreme gradient boosting (XGB), and random forest (RF)8. We tuned the hyperparameters (maximum depth, learning rate, and number of leaves) with a Bayesian search mechanism9 and 5-fold cross-validation. We conducted 5-fold cross-validation to evaluate model performance using the F1-score to account for imbalanced protocol selections. SHAP (SHapley Additive exPlanation) values were plotted to visualize relative feature contributions to final predictions. The tuned models were then used to predict protocols and report their probabilities. This pipeline produces a list of the top-three radiology protocol suggestion list, with the probability of each protocol. The accuracy of the selections was evaluated based on F1-scores.
Results and Discussion
Cross-validation yielded a mean F1-score of 78.5, 75.6, and 72.6% for the XGB, LGBM, and RF models, respectively, on the training and validation sets (Fig. 2). According to our SHAP plots, the key features contributing to protocol selection were radiology title (description of imaging order requested by ordering physician), International Classification of Diseases (ICD-10) code, and sex. Expectedly, sex is an important variable for some protocols such as Prostate Local. These features were significant determinant factors in RF, LGBM, and XGB models (Fig. 3). The most probable predicted protocols were reported for radiologist review with their probabilities, as shown in Fig. 4. We obtained accumulated F1 score of 95.6, 91.0, and 90.4% for the XGB, LGBM, and RF models, respectively, for the top-three predicted protocols on the test set (Fig. 5), indicating that the ML-selected recommendations would be useful for protocol selection in the context of a clinical decision support system. The system could also alert radiologists to potential radiology title-protocol mismatches.Conclusions
ML models trained on EMR data can generate appropriate MRI protocol suggestions and identify the most important features in determining protocol suitability. Cross-validation-generated top-one F1 scores exceeded 75% for the present ML system, indicating that its outputs provide guidance toward highly suitable protocol selections. The top-three selections recommended (F1 scores 90%) can be used to improve imaging protocol precision (meaning more tightly coupling the exam protocol to the clinical indication), which can help to reduce costs and support patient outcomes. Our decision-tree based models is currently being incorporated into a decision support system to facilitate accurate MRI protocoling in routine practice by presenting the top three protocol recommendations.Acknowledgements
This work has been supported and funded by General Electric (GE) Healthcare.References
- Stewart C, MRI units per million: by country 2019, Statista. Oct 26, 2021:A1. https://www.statista.com/statistics/282401/density-of-magnetic-resonance-imaging-units-by-country/, Accessed November 4, 2022.
- Brown A, Marotta T. A natural language processing-based model to automate MRI brain protocol selection and prioritization. Acad. Radiol. 2017;24(2):160-166.
- Kalra A, Chakraborty A, Fine B, et al. Machine learning for automation of radiology protocols for quality and efficiency improvement. J. Am. Coll. Radiol. 2020;17(9):1149-1158.
- Abuzaid MM, Tekin HO, Reza M, et al. Assessment of MRI technologists in acceptance and willingness to integrate artificial intelligence into practice. Radiog. 2021;27:S83-S87.
- Linna N, Kahn JrCE. Applications of natural language processing in radiology: a systematic review. Int. J. Med. Inform. 2022;104779.
- Donnelly LF, Grzeszczuk R, Guimaraes CV. Use of natural language processing (NLP) in evaluation of radiology reports: an update on applications and technology advances. Semin. Ultras. CT MRI. 2022;43(2):176-181.
- Ferreira A, Figueiredo M. Boosting algorithms: A review of methods, theory, and applications. Ens. Mach. Learn., Springer, Boston, MA, USA; 2012.
- Klug M, Barash Y, Bechler S, et al. A gradient boosting machine learning model for predicting early mortality in the emergency department triage: devising a nine-point triage score. J. Gen. Intern. Med. 2020;35(1): 220-227.
- Owen L. Hyperparameter tuning with python. 2nd Ed. Birmingham: Packt Pub., O'Reilly. 2022: 133-136. https://learning.oreilly.com/library/view/hyperparameter-tuning-with/9781803235875/, Accessed November 7, 2022.
Figures
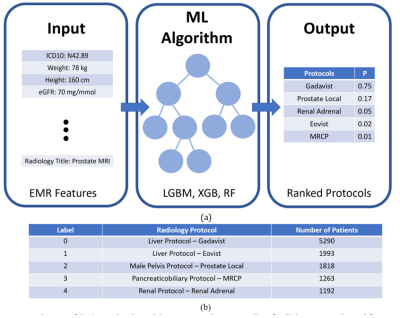
Fig. 1 a)
Diagram of the input signal, model structure, and output, b) list of radiology protocols used for MRI scanning and the
number of patients for the top 5 protocols in the original dataset. The number
of patients and their corresponding exam orders in the entire original dataset (including
all protocols) was 19,590 and 38,010, respectively, and it was reduced to 6,882
and 11,251 after selecting the top 5 protocols and performing further
preprocessing methods.
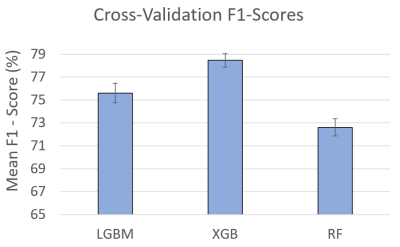
Fig. 2 Mean F1-score for predicting
radiology protocols with 5-fold cross-validation. The results indicate
obtaining the acceptable range (>70%) that confirms
the capability of the proposed pipeline to predict new data, avoid overfitting,
and generalize to an independent dataset. It should be noted that
cross-validation was applied to the training and validation sets (75% of the
data) to keep the test set intact for the radiology protocol predictions.
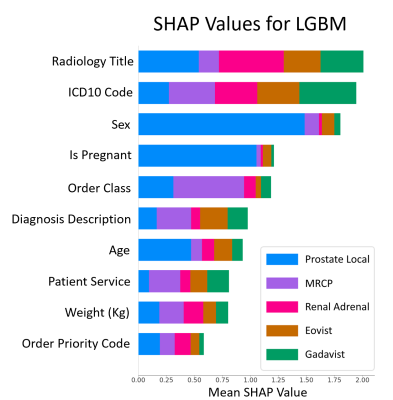
Fig.
3 LGBM SHAP plot illustrating the contribution of each
feature to the prediction. The plot indicates that Radiology Title (i.e.,
ordered exam imaging title), International Classification of Diseases Code
(ICD10), and Sex are the most significant features. Order Class indicates if
the imaging is performed at clinic/hospital/external. Patient Service indicates
the type of service the patient received (e.g., critical care, emergency).
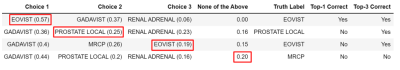
Fig. 4 Four examples of the top three suggested radiology
protocols with their probabilities according to the XGB model. The red boxes
indicate appropriately selected protocol. Top-1/Top-3 correct columns indicate
whether the selected protocol was among the top one/top three suggested protocols.
In case 1, Choice 1 was correct; however, two more choices were provided for
using this pipeline as a clinical decision support system.
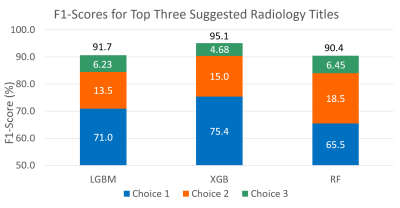
Fig. 5 F1-scores for the three top selected radiology protocols on
the held-out test set of five protocols for 1,885 patients. The results (≥ 90%)
confirms the appropriate performance of the proposed clinical decision support
system in recommending top-three radiology protocols.
DOI: https://doi.org/10.58530/2023/3702