3686
Optimizing MRSI semi-LASER for mouse brain tumor studies: evaluation of increased spatial resolution and k-space sampling strategies1Departament de Bioquímica i Biologia Molecular, Universitat Autònoma de Barcelona, Cerdanyola del Valles, Spain, 2Servei de Ressonància Magnètica Nuclear, Universitat Autonoma de Barcelona, Cerdanyola del Valles, Spain, 3Department of Imaging Physics,, The University of Texas M.D. Anderson Cancer Center, Houston, TX, United States, 4Centro de Investigación Biomédica en Red en Bioingeniería, Biomateriales y Nanomedicina (CIBER-BBN), Cerdanyola del Valles, Spain, 5Institut de Biotecnologia i de Biomedicina (IBB), Universitat Autònoma de Barcelona, Cerdanyola del Valles, Spain
Synopsis
Keywords: Spectroscopy, Cancer, Brain Tumor
MRSI-based nosological images have been previously applied to monitor therapy response in a murine model of GL261 glioblastoma using a commercially available MRSI-PRESS sequence. Due to the heterogeneous nature of glioblastomas, increasing spatial resolution could provide additional insight into changes related to therapy response. The in-house implementation of the MRSI-semi-LASER sequence, using weighted acquisition, has allowed us a 20% slice thickness reduction without losing relevant spectral quality. Moreover, different k-space sampling strategies have been evaluated showing significant SNR increase with elliptical sampling, which may allow further increase in the spatial resolution or reduction in the total experimental time.INTRODUCTION
Magnetic resonance spectroscopic imaging (MRSI) provides information on biochemical environment of brain tumors, in addition to anatomic information by imaging techniques [1]. We have been applying pattern recognition approaches to MRSI data to assess therapy response in a murine model of GL261 glioblastoma (GB) [2]. The multi-slice 1H MRSI protocol used consisted of acquiring three to four consecutives grids across tumors, of 1 mm slice thickness, using a commercially available MRSI-PRESS sequence. Since GBs display intratumoral heterogeneity, higher spatial resolution may be of interest for improved tumor evaluation. In this respect, the in-house implemented preclinical MRSI-semi-LASER sequence has proven to be suitable for high resolution multi-voxel spectroscopic acquisitions in mouse brain studies [3]. MRSI-semi-LASER data have shown improved spatial distribution homogeneity and higher signal to noise ratio (SNR) as compared to the conventional MRSI-PRESS sequence. Our purpose was to evaluate spectral quality of MRSI-semi-LASER data sets, acquired at 0.8mm slice thickness, in GL261 tumor-bearing mice. Moreover, different k-space sampling schemes were also evaluated.METHODS
Experiments were performed on a 7T Bruker Biospec 70/30 USR preclinical scanner using ParaVision 5.1 (Bruker BioSpin GmbH, Ettlingen, Germany) equipped with a mini-imaging gradient set (400 mT/m), a 72-mm inner diameter linear volume coil used as transmitter and a dedicated mouse brain surface coil as receiver. C57BL/6 female mice were obtained from Charles River Laboratories and housed in the animal facility of the Universitat Autònoma de Barcelona where GL261 GB tumors were generated. MRSI-semi-LASER acquisition parameters: TE, 27 ms; TR, 2500 ms; VAPOR water suppression; FOV, 17.6 x 17.6 mm2; slice thickness, 1 mm or 0.8 mm; voxel size, 6.6 x 6.6 mm2 (middle grids) and 5.5 x 5.5 mm2 (outer grids); acquisition matrix size of 8x8 interpolated to 32 x 32, thus resulting in a voxel matrix size of 12 x 12 and 10 x 10 respectively in middle and outer grids; spectral width, 4006.41 Hz; number of acquisitions, 512 (standard and weighted), 520 (elliptical); total acquisition time, 21 min per grid. SNR values were estimated as maximum signal intensity divided by standard deviation of the noise.RESULTS
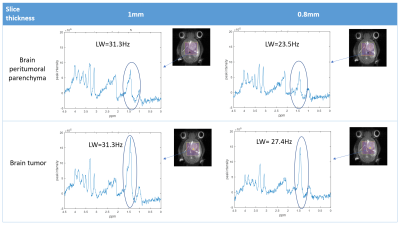
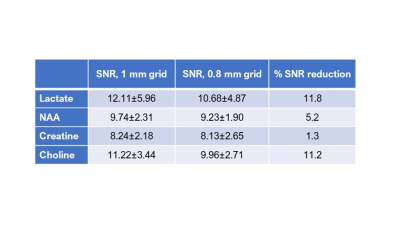
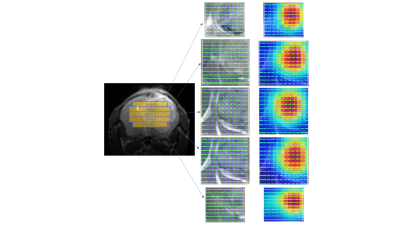
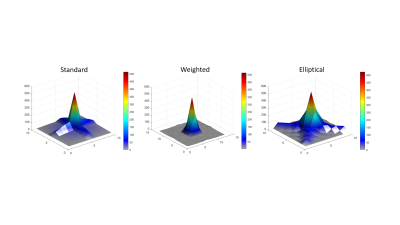
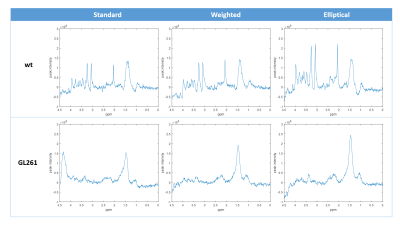
Comparison of 1 mm versus 0.8 mm slice thickness MRSI-semi-LASER with weighted k-space acquisitions is shown in Figure 1, where representative spectra from brain peritumoral parenchyma and tumor regions are displayed. Table in Figure 2 shows mean SNR values of the whole grid for several metabolites, reflecting less than the expected 20% SNR decrease in the 0.8 mm slice grid compared to 1 mm. Five consecutive 0.8 mm slice thickness grids were able to be satisfactorily acquired individually across the tumor in GL261 tumor-bearing mice (Figure 3). These five slices covered the same volume as the four 1mm slices that would be acquired with our previous protocol. An evaluation of the different k-space acquisition schemes was performed in a phantom, in wild-type mouse brain, and finally in GL261 tumor-bearing mice. The point spread functions (PSF) for standard, weighted and elliptical k-space modes are shown in Figure 4, revealing strong suppression of the side lobes with the weighted scheme, which was the scheme used by default in our previous studies. Figure 5 shows representative MRSI-semi-LASER spectra from a wild-type mouse brain parenchyma and a tumor-bearing mouse comparing the three k-space acquisition schemes. MRSI-semi-LASER spectra acquired with elliptical k-space demonstrated higher SNR compared to standard and weighted acquisition modes. Mean SNRs for the NAA peak in normal brain parenchyma were calculated to be 12.20 ± 1.75 for standard, 13.18 ± 1.69 for weighted, and 15.38 ± 2.34 for elliptical acquisition modes. Moreover, elliptical phase encoding spectra showed comparably good spectrum quality, with no evidence of spurious signals.DISCUSSION
Increased spatial resolution was achieved using MRSI-semi-LASER in GL261 tumor-bearing mice, being able to acquire an extra slice within the same tumor volume. When decreasing the slice thickness from 1 mm to 0.8 mm, we expected to see a 20 % reduction in SNR, however, SNR was maintained above the expected level (Figure 2). Narrower water peak widths at half height were observed in spectra acquired at 0.8 mm indicating better B0 homogeneity in thinner slices, which results in narrower and more intense peaks with SNR larger than predicted based on volume differences. With elliptical phase encoding, where corners of k-space are skipped, a higher number of phase encoding steps in each direction can be acquired within the same acquisition time, as compared to standard or weighted acquisition. This causes broadening of the spatial response function but also increases the SNR. The weighted mode had been used by default in our previous studies because its known improved PSF as compared to standard or elliptical acquisition modes. Nonetheless, elliptical acquisition provided good quality spectra with no apparent artefactual signals and a 16.7 % SNR increase with respect to weighted acquisitions.CONCLUSION
In this study we have shown the added value of fully adiabatic localization in MRSI-semi-LASER. Moreover, the increased SNR obtained with elliptical phase encoding may allow to further increase the spatial resolution or reduce the experimental time in order to improve pattern spectral analysis in our murine brain tumor models.Acknowledgements
This work is funded by the INSPiRE-MED project 2019-2022 (H2020/MCSA COFUND, Grant agreement ID: 813120) and PID2020-113058GB-I00 (MCIN/AEI/10.13039/501100011033).References
[1] Horská A. and Barker P. B. Imaging of Brain Tumors: MR Spectroscopy and Metabolic Imaging. Neuroimaging Clin N Am. 2010 20(3): 293–310. doi: 10.1016/j.nic.2010.04.003
[2] Arias-Ramos, N.; Ferrer-Font, L.; Lope-Piedrafita, S.; Mocioiu, V.; Julià-Sapé, M.; Pumarola, M.; Arús, C.; Candiota, A.P. Metabolomics of Therapy Response in Preclinical Glioblastoma: A Multi-Slice MRSI-Based Volumetric Analysis for Noninvasive Assessment of Temozolomide Treatment. Metabolites 2017, 7, 20. https://doi.org/10.3390/metabo7020020
[3] Javed Z, Martinez G, Candiota AP, Cabanas M, Lope-Piedrafita S. High resolution multi-voxel spectroscopy using CSI-semi-LASER for mouse brain preclinical studies. ISMRM 2022 (abstract 2329)
Figures