3684
Test-retest reliability of PRESS- and sLASER-localized multi-metabolite spectral editing1Department of Radiology, Weill Cornell Medicine, New York, NY, United States, 2GE Healthcare, Berlin, Germany
Synopsis
Keywords: Spectroscopy, Spectroscopy, Reproducibility
Multi-metabolite spectral-edited MRS enables the efficient detection of multiple low-concentration metabolites. Two such approaches, HERMES and HERCULES, directly edit two or more metabolites in a single scan. However, the reproducibility of these techniques has yet to be fully established. This study investigated PRESS- and sLASER-localized HERMES and HERCULES test-retest reliability, with the additional aim of demonstrating that sLASER localization is the better of the two sequences for obtaining reproducible measurements. Overall, the test-retest reliability of sLASER was found to be higher than that of PRESS.Introduction
Spectral editing is the most used MRS approach for in vivo measurement of low-concentration metabolites such as γ-aminobutyric acid (GABA) and glutathione (GSH). However, a disadvantage is that typically only a single metabolite is targeted in each acquisition. In addition to the relatively long scan times required to improve SNR, single-metabolite editing can be inefficient when multiple low-concentration metabolites may be of interest. Recently, novel editing methods, including HERMES1,2 and HERCULES3, were introduced where multiple scalar-coupled metabolites can be targeted in a single acquisition without increasing total scan time. This is achieved through multiplexed editing schemes (with four or more steps) and Hadamard reconstruction. In this study, the test-retest reliability of multi-metabolite editing using sLASER for volume localization was compared to that of standard PRESS localization.Methods
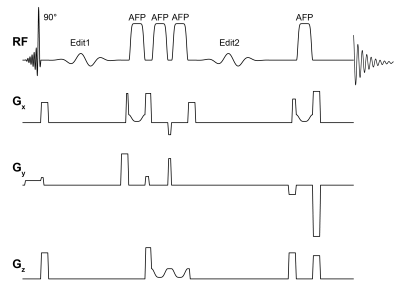
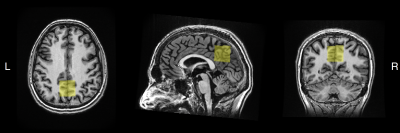
Data acquisitionAll data were collected on a 3T GE Discovery MR750 MRI scanner using a 32-channel RF head coil for receive and a body coil for transmit. Thirteen adult volunteers (F/M = 8/5; mean ± 1 std. = 36.7 ± 18.5 years) were scanned twice in two sessions separated by a time delay (median interval between scans = 0 days; range = 0–29 days). High-resolution T1-weighted BRAVO structural scans (FSPGR; TE/TR/TI = 5.2/12.2/725 ms; voxel resolution = 0.9 × 0.9 × 1.5 mm3; matrix size = 256 × 256) were first acquired for accurate voxel placement. MRS data were then acquired in the following order (and were not counterbalanced between sessions): (1) HERMES-PRESS; (2) HERMES-sLASER; (3) HERCULES-PRESS; (4) HERCULES-sLASER. The pulse sequence diagram for sLASER-localized multi-metabolite editing is shown in Figure 1. The following acquisition parameters were used for all the MRS sequences: TE/TR = 82/2000 ms; 5000 Hz spectral width; 4096 spectral points; 224 transients; voxel resolution = 3 × 3 × 3 cm3. The MRS voxel was placed in the medial parietal lobe (Figure 2). PRESS-localized scans used CHESS for water suppression, while sLASER scans used VAPOR. Unsuppressed water reference scans were also collected for signal referencing.
Data analysis
Spectra were processed using Osprey4 (v2.3.0) and involved the following steps: (1) coil combination using generalized least squares5; (2) eddy-current correction; (3) robust spectral registration6; (4) signal averaging; (5) residual water filtering using HSVD; and (6) reconstruction of four Hadamard-combination subspectra based on the four subspectra, labeled A, B, C, and D.
Reconstructed spectra, denoted DIFF1 (A+B–C–D) and DIFF2 (A–B+C–D), were fitted using nonlinear least-squares linear-combination modeling. Basis sets were created using high spatial resolution (101 × 101 points) density-matrix numerical simulations. These were run using the HERMES- and HERCULES-edited PRESS and sLASER pulse sequence parameters to simulate metabolite signal lineshapes accurately. Simulations were run in MRSCloud7 using a novel 1D projection method8 and coherence pathway filtering9 to reduce computation time. Included metabolites were Asc, Asp, Cr, GABA, GPC, GSH, Gln, Glu, Gly, Lac, mI, NAA, NAAG, PCh, PCr, PE, Ser, sI, and Tau. Macromolecule and lipid resonances were parameterized using Gaussian functions.
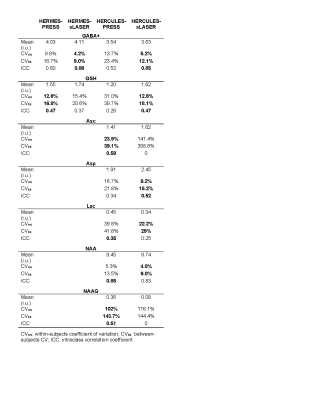
SNR and linewidth were calculated based on the Cr signal in the SUM (A+B+C+D) spectrum to assess data quality. Metabolite levels were quantified relative to unsuppressed water and are reported in institutional units (i.u.). Partial-volume tissue correction was not performed. Metabolites of interest were Asc, Asp, GABA+ (with co-edited macromolecules), GSH, Lac, NAA, and NAAG. Within- and between-subjects coefficients of variation (CVs) and intraclass correlation coefficients (ICCs) (two-way mixed-effects models of absolute agreement) were calculated in R and used to assess reliability.
Results
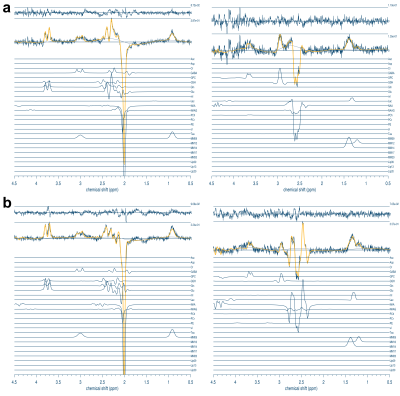
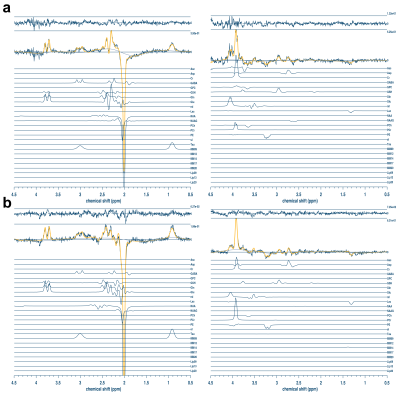
All spectra were visually inspected for signal artifacts. Sample HERMES-PRESS and -sLASER DIFF1 and DIFF2 spectra are shown in Figure 3, while sample HERCULES-PRESS and -sLASER DIFF1 and DIFF2 spectra are shown in Figure 4. The sLASER spectra had less signal distortion, particularly in the 4–4.5 ppm range where artifactual outer-volume echoes typically appear.Average Cr SNR was 197 ± 26, 198 ± 21, 193 ± 25, and 200 ± 24 for HERCULES-PRESS, -sLASER, HERMES-PRESS, and -sLASER, respectively. The average Cr linewidth was 7.5 ± 1.1 Hz, 6.9 ± 0.7 Hz, 7.5 ± 0.9 Hz, and 6.8 ± 0.7 Hz for the four experiments, respectively. Reproducibility statistics are shown in Table 1. Overall, the sLASER-localized acquisitions showed better reproducibility. However, for Asc and NAAG, the CVs were very high for the sLASER implementation, indicating poor reproducibility. Additionally, the reproducibility of GSH measurements for HERMES-sLASER was lower than that for HERMES-PRESS for reasons that are unclear.
Conclusion
Overall, sLASER localization showed higher reproducibility than PRESS localization for multi-metabolite editing. However, there is room for improvement. Specifically, the editing scheme used for HERCULES was initially derived to edit as many scalar-coupled metabolites as is seemingly allowable to preserve the orthogonality of the four-step editing experiment of HERCULES. Alternative editing schemes need to be explored where a subset of metabolites is targeted for optimal signal yield and reproducibility for a given research question.Acknowledgements
This work was supported by NIH grant K99 EB028828.References
1. Chan KL, Puts NAJ, Schär M, Barker PB, Edden RAE. HERMES: Hadamard encoding and reconstruction of MEGA-edited spectroscopy. Magn Reson Med. 2016;76(1):11-19. doi:10.1002/mrm.26233
2. Saleh MG, Oeltzschner G, Chan KL, et al. Simultaneous edited MRS of GABA and glutathione. Neuroimage. Nov 15 2016;142:576-582. doi:10.1016/j.neuroimage.2016.07.056
3. Oeltzschner G, Saleh MG, Rimbault D, et al. Advanced Hadamard-encoded editing of seven low-concentration brain metabolites: Principles of HERCULES. Neuroimage. Jan 15 2019;185:181-190. doi:10.1016/j.neuroimage.2018.10.002
4. Oeltzschner G, Zollner HJ, Hui SCN, et al. Osprey: Open-source processing, reconstruction & estimation of magnetic resonance spectroscopy data. J Neurosci Methods. Sep 1 2020;343:108827. doi:10.1016/j.jneumeth.2020.108827
5. An L, Willem van der Veen J, Li S, Thomasson DM, Shen J. Combination of multichannel single-voxel MRS signals using generalized least squares. J Magn Reson Imaging. Jun 2013;37(6):1445-50. doi:10.1002/jmri.23941
6. Mikkelsen M, Tapper S, Near J, Mostofsky SH, Puts NAJ, Edden RAE. Correcting frequency and phase offsets in MRS data using robust spectral registration. NMR Biomed. 2020;33(10):e4368-e4368. doi:10.1002/nbm.4368
7. Hui SCN, Saleh MG, Zollner HJ, et al. MRSCloud: A cloud-based MRS tool for basis set simulation. Magn Reson Med. Jul 1 2022;(June):1-11. doi:10.1002/mrm.29370
8. Zhang Y, An L, Shen J. Fast computation of full density matrix of multispin systems for spatially localized in vivo magnetic resonance spectroscopy. Med Phys. Aug 2017;44(8):4169-4178. doi:10.1002/mp.12375
9. Landheer K, Swanberg KM, Juchem C. Magnetic resonance Spectrum simulator (MARSS), a novel software package for fast and computationally efficient basis set simulation. NMR Biomed. May 2021;34(5):e4129. doi:10.1002/nbm.4129
Figures