3683
Selective Extraction of J-coupled Signals Using MEGA-PRESS with Conditional Saturation Pulses
Hidenori Takeshima1, Shuki Maruyama2, and Masao Yui3
1Imaging Modality Group, Advanced Technology Research Department, Research and Development Center, Canon Medical Systems Corporation, Kanagawa, Japan, 2Imaging Modality Group, Advanced Technology Research Department, Research and Development Center, Canon Medical Systems Corporation, Tochigi, Japan, 3Research and Development Center, Canon Medical Systems Corporation, Tochigi, Japan
1Imaging Modality Group, Advanced Technology Research Department, Research and Development Center, Canon Medical Systems Corporation, Kanagawa, Japan, 2Imaging Modality Group, Advanced Technology Research Department, Research and Development Center, Canon Medical Systems Corporation, Tochigi, Japan, 3Research and Development Center, Canon Medical Systems Corporation, Tochigi, Japan
Synopsis
Keywords: Pulse Sequence Design, Spectroscopy
In this abstract, a spectral editing method using MEGA-PRESS with conditional frequency-selective saturation pulses is proposed. The proposed method used additional frequency-selective saturation blocks before the excitation block. The saturation pulses were activated only when the MEGA pulses were inactive. When saturation pulses were activated, the sequence acted as the conventional MEGA-PRESS whose editing pulses were inactive except that the passband of the saturation pulse was not excited. Spectrum reconstruction for the proposed method was same as that for the conventional MEGA-PRESS. Experimental results showed that the proposed method selectively extracted J-coupled signals with all frequencies.Introduction
For edited MR spectroscopy1, sequences with frequency-editing refocusing pulses such as band selective inversion with gradient dephasing (BASING)2 and Mescher-Garwood (MEGA)3 pulses are used commonly for analyzing J-coupled evolution. These sequences acquire differences between signals with editing pulses and signals without the editing pulses. For non-editing frequency bands, J-coupled evolution can be selectively observed. However, in the editing frequency band, J-coupled evolution cannot be analyzed easily since non-J-coupled signals such as singlets are overlapped with J-coupled signals. Such signals are not observable when other editing techniques such as a double quantum filter4 are used.In this abstract, a spectral editing method using 1H MEGA- Point RESolved Spectroscopy (PRESS) with conditional frequency-selective saturation pulses is proposed. Similar to other editing techniques, the proposed method selectively extracted J-coupled signals with all frequencies.
Methods
Pulse sequenceAs shown in Figure 1, the proposed method used additional frequency-selective saturation blocks before the excitation block of PRESS. The saturation pulses were centered with the editing frequency (referred to as active) only when the MEGA pulses were centered with a non-editing frequency (referred to as inactive). When saturation pulses were activated (named Sat), the sequence acted as MEGA-PRESS whose editing pulses were inactive except that the passband of the saturation pulse was not excited. When the MEGA pulses were activated (named Nosat), the sequence acted as MEGA-PRESS whose editing blocks were used. For both the saturation and the MEGA pulses, a Gaussian pulse with duration of 21.2 milliseconds was used. A block of VAriable Power and Optimized Relaxations delays (VAPOR)5 was used before the excitation block for water suppression.
To extract J-coupled signals selectively, the signals of Nosat were subtracted from the signals of Sat. The evolution of unsaturated spins and J-coupling was active in the Sat cases. In the Nosat cases, the evolution of all spins and J-coupling was active and the evolution of J-coupling in the passband of the MEGA pulses was spoiled in the MEGA blocks.
In the following evaluations, the editing frequency was adjusted to approximately 1.9 ppm using estimated frequency of water. The editing frequency for inactive cases was determined for simultaneous water suppression of MEGA-PRESS.
Evaluations using phantoms
Two phantoms named Phantom #1 and Phantom #2 were scanned using a 3-T scanner and a 32-channel receiver head coil with NEX of $$$64 \times 2$$$. Other scan parameters were TE of 68 milliseconds, TR of 2000 milliseconds, dwell time of 384 microseconds, samples per readout of 1024, and volume-of-interest (VOI) of $$$3 \times 3 \times 3$$$ cm3. In addition to the subtraction of the Nosat spectrum from the Sat spectrum, individual spectra were also reconstructed for Phantom #1.
The concentrations of the metabolites used in these phantoms are shown in Figure 2. Phantom #1 simulated the spectra of a normal brain6 with an inhibitory neurotransmitter gamma-aminobutyric acid (GABA). Phantom #2 simulated isocitrate dehydrogenase (IDH)-mutant brain tumor tissue7 with oncometabolite 2-hydroxyglutarate (2-HG). Spectra of GABA and 2-HG have measurable spins with J-couplings at approximately 3.0 ppm and 4.0 ppm, respectively, when editing pulses are applied to 1.9 ppm 1. These spectra also have measurable spins in 2.2 - 2.3 ppm for the same editing pulses. In addition, N-acetylaspartate (NAA) has a singlet at 2.0 ppm near editing frequency. These phantoms also contained several other metabolites including NAA as shown in Figure 2.
Evaluation using volunteer data
A healthy volunteer was scanned using another 3-T scanner and a 12-channel receiver head neck coil with NEX of $$$128 \times 2$$$. Other scan parameters were same as those for the phantoms. The subtracted spectra were reconstructed for the volunteer data.
Results
Individual spectra of Phantom #1 are shown in Figure 3. It could be confirmed that the unsuppressed spectrum of MEGA off was suppressed in the spectrum of Sat near the editing frequency at 1.9 ppm.The subtracted spectra of Phantom #1 and Phantom #2 are shown in Figure 4. The editing-related spectra were found at 3.0 ppm in Phantom #1 and at 4.0 ppm in Phantom #2. In addition, unlike the spectrum of MEGA, the spectrum of the proposed method clearly suppressed the singlet of NAA at 2.0 ppm.
The spectra of the volunteer are shown in Figure 5. The editing-related spectra were detectable at 3.0 ppm. Similar to the phantom cases, the spectrum of the proposed method revealed J-coupled signals near the editing frequency at 1.9 ppm.
Discussion
The results showed that the singlet of NAA at 2.0 ppm was selectively suppressed in the spectra of the proposed method. These results implied that the proposed method could reveal J-coupled signals around 1.9 ppm. While it could not be confirmed clearly, spectra come from macromolecule (MM)8 seemed to be suppressed except but signals coupled within the editing frequency band.Conclusion
A spectral editing method using MEGA-PRESS with conditional frequency-selective saturation pulses has been proposed. The proposed method used additional frequency-selective saturation blocks before the excitation block of PRESS. The obtained spectra implied that the information of J-coupled signals could be used as additional information for analyzing spectra.Acknowledgements
No acknowledgement found.References
1. Harris AD, Saleh MG, Edden RA. Edited 1 H magnetic resonance spectroscopy in vivo: Methods and metabolites. Magn Reson Med 2017; 77(4):1377-1389.2. Star-Lack J, Nelson SJ, Kurhanewicz J, Huang LR, Vigneron DB. Improved water and lipid suppression for 3D PRESS CSI using RF band selective inversion with gradient dephasing (BASING). Magn Reson Med 1997; 38(2):311-321.
3. Mescher M, Merkle H, Kirsch J, Garwood M, Gruetter R. Simultaneous in vivo spectral editing and water suppression. NMR Biomed 1998; 11(6):266-272.
4. McLean MA, Busza AL, Wald LL, Simister RJ, Barker GJ, Williams SR. In vivo GABA+ measurement at 1.5T using a PRESS-localized double quantum filter. Magn Reson Med 2002; 48(2):233-241.
5. Tkác I, Starcuk Z, Choi IY, Gruetter R. In vivo 1H NMR spectroscopy of rat brain at 1 ms echo time. Magn Reson Med. 1999; 41(4): 649-656.
6. https://www.goldstandardphantoms.com/products/spectre/
7. Nagashima H, Tanaka K, Sasayama T, et al. Diagnostic value of glutamate with 2-hydroxyglutarate in magnetic resonance spectroscopy for IDH1 mutant glioma. Neuro Oncol. 2016; 18(11): 1559-1568.
8. Behar KL, Rothman DL, Spencer DD, Petroff OA. Analysis of macromolecule resonances in 1H NMR spectra of human brain. Magn Reson Med 1994; 32(3):294-302.
Figures
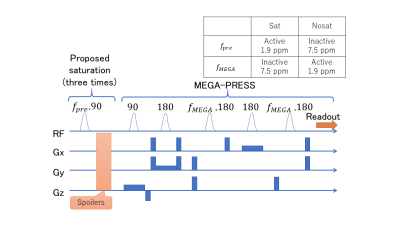
The pulse sequence of the proposed method. A frequency-selective saturation blocks was added before the excitation block of PRESS. The saturation pulses were activated only when the MEGA pulses were inactive. The saturation pulses were used three times and were inserted in waiting times of a VAPOR block. The VAPOR block is omitted for conciseness. The table in this figure represents the editing frequencies used for the water at approximately 4.7 ppm.
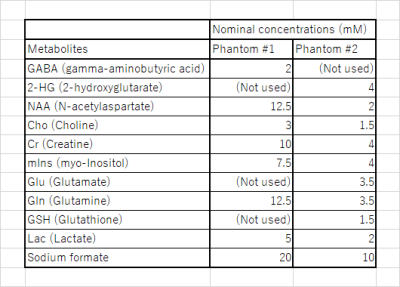
Details of phantoms used in experiments. Phantoms #1 and #2 contained GABA and 2-HG, respectively. Both phantoms were placed in 500-milliliter round-bottom glass flasks. As their solvents, phosphate-buffered saline (PBS) solutions were used.
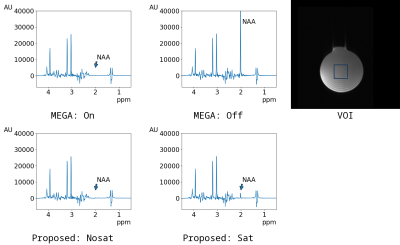
Individual spectra of the phantom #1. The spectrum
of MEGA on was almost same as that of Nosat. However, the unsuppressed spectrum
of MEGA off was suppressed in the spectrum of Sat near the editing frequency at
1.9 ppm.
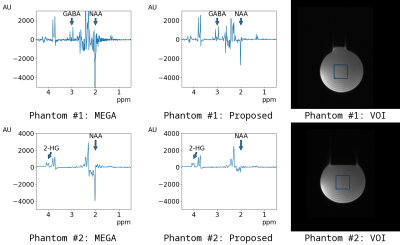
Subtracted spectra of Phantom #1 and Phantom #2. For all phantoms, editing-related spectra were detectable at 3.0 ppm in Phantom #1 and at 4.0 ppm in Phantom #2. In addition, unlike the spectrum of MEGA, the spectrum of the proposed method clearly suppressed the singlet of N-acetylaspartate (NAA) at 2.0 ppm. J-coupled signals were not suppressed near the editing frequency at 1.9 ppm.

Spectra of the volunteer. Editing-related spectra were detectable at 3.0 ppm. Similar to the phantom cases, the spectrum of the proposed method clearly suppressed the singlet of N-acetylaspartate (NAA) at 2.0 ppm.
DOI: https://doi.org/10.58530/2023/3683