3657
Deformation-ENcoding Transformer (DENT) for High Frame Rate for Phase Contrast MRI
Manuel A Morales1, Amine Amyar1, Siyeop Yoon1, Jennifer Rodriguez1, Martin S Maron2, Ethan J Rowin2, Shiro Nakamori1, Jiwon Kim3, Robert M Judd4, Jonathan W Weinsaft3, Warren J Manning1, and Reza Nezafat1
1BIDMC, Boston, MA, United States, 2Tufts Medical Center, Boston, MA, United States, 3Weill Cornell Medicine, New York, NY, United States, 4Duke University, Durham, NC, United States
1BIDMC, Boston, MA, United States, 2Tufts Medical Center, Boston, MA, United States, 3Weill Cornell Medicine, New York, NY, United States, 4Duke University, Durham, NC, United States
Synopsis
Keywords: Flow, Cardiovascular
Quantification of blood flow using 2D or 4D phase-contrast (PC) MRI is routinely being used to evaluate blood flow in cardiovascular disease. However, PC has only a modest temporal resolution compared to echocardiography. We sought to develop a deformation-encoding transformer (DENT) model for cardiac frame interpolation and evaluate its potential in increasing temporal resolution. DENT was trained using a large multi-center (centers = 3), multi-vendor (vendors = 3) and multi-field strength (1.5T, 3T) cine MRI dataset (patients = 3178). The model was successfully applied to 2D/4D-PC MRI without modifications, enabling a 2-fold gain in temporal resolution.Background
Quantification of blood flow using 2D or 4D phase-contrast (PC) MRI is routinely being used to evaluate blood flow in cardiovascular disease. Cardiac 2D or 4D phase contrast (PC) flow imaging allows evaluation of blood flow and hemodynamics within cardiovascular system. In ECG-segmented 2D or 4D PC imaging, there is a trade-off between scan time and spatiotemporal resolution. Insufficient temporal resolution leads to imprecise peak velocity measurements. We sought to develop a Deformation ENcoding Transformer (DENT) for high-frame-rate 2D/4D PC MRI.Methods
DENT worked in the image domain and generated forward-backwards mappings describing the underlying deformation of multiple cardiac phases. Such deformation, as well as a blending mask, was used to synthesize images with high framerate. Input was T = 4 cine images of size W × H collected with temporal resolution ∆t at t - ∆t, t, t + ∆t and t + 2∆t. Output was interpolated image at t + 0.5∆t (Fig. 1a). An embedding layer extracted F = 32 features per pixel from inputs. Downsampling layers reduced image dimensionality by half while doubling feature dimensionality. Inputs to transformer layers were split into windows. Multiscale attention was used to learn a spatiotemporal correspondence within each window (Fig. 1b). For spatial attention, inputs were split into N = W•H•T / M2 windows of size M2 × F along spatial dimension (M = 8). For temporal attention, inputs were split into N = W•H windows of size T × F (Fig. 1c). Decoder layers upsampled the dimensionality of encoder outputs by 2 while reducing feature dimensionality by half. These were passed to the motion synthesizer block. Additional convolutional layers in the block upsampled the input. The final outputs of the model were deformation , scaling components , and blending mask (Fig. 1d). A new cardiac frame was synthesized from each input image using deformation and scaling components via bilinear interpolation. The blending mask was used to combine synthesized images onto a single frame at t + 0.5∆t. Multi-center (centers = 3), multi-vendor (GE, Philips, Siemens), multi-field strength (1.5T, 3T) scans from 3178 patients (2139 male, 54 ± 16 years) undergoing clinical MRI for various cardiac indications were used for training. Cine images were collected using a breath-hold ECG-gated segmented SSFP at 1.5T (n = 1831) and 3T (n = 1347) in short axis and 2-, 3- and 4-chamber. A sample was as a cine slice with T ≥ 7 frames; an epoch one optimization loop across all training samples. First, a center-frame was randomly selected and used as the ground-truth. Second, 4 frames adjacent to were selected as inputs: either or , which had a 2∆t and 4∆t temporal resolution, accordingly (Fig. 2). Each cine frame was normalized by min-max prior to processing. Ground-truth and input images were randomly cropped to 256 × 256. Training used a batch size = 10 for 100 epochs (~200 hours) using AdaMax. Learning rate was 2 × 10-2 and gradually decayed to 1 × 10-6. 2D or 4D PC MRI imaging dataset from patients who were imaged using Siemens 3T Vida system were extracted to demonstrate feasibility of DENT-enabled high-frame-rate flow. Breath-hold ECG-segmented 2D PC scans were acquired with the following imaging parameters: spatial resolution = 1.9 × 1.9 mm2, temporal resolution = 37 ms; GRAPPA rate = 3, number of phase-encode line per segment = 4. Navigated free-breathing ECG-segmented 4D PC scans were acquired (acquisition time = 4.5 min) with the following imaging parameters: TE/TR = 2.3/4.58 ms, spatial resolution = 2.5 × 2.5 × 2.5 mm3, acquired temporal resolution = 37 ms, reconstructed number of phases = 25 ms, vendor-provided WIP compressed sensing rate 7.2, number of phase-encode line per segment = 2. Encodings , and were derived from the anatomical images of flow datasets (Fig. 3). These encodings were applied to both the anatomical and flow images to synthesize a new cardiac frame. For 4D flow, the encodings were applied to flow images in x, y and z directions separately. 4D flow were processed and visualized in Medis Suite (4.0.50.2).Results
trained DENT model enabled both anatomical and flow images with high-frame-rate, as demonstrated for 2D PC (Fig. 4). A two-fold gain was achieved in 4D flow datasets with simulated and reconstructed temporal resolutions of 54 and 27 ms, accordingly. Our approached resulted in 4D flow with temporal resolution as low as 14 ms (Fig. 5). DENT was trained in large cine-based dataset. Translational feasibility for PC was demonstrated. However, additional (i.e., phantom) studies should be performed to assess the contribution of each encoding component. For instance, in cases where deformation does not play a major role (e.g., center of vessels), the synthesized flow values are essentially a linear combination of the four adjacent frames. The coefficients of that linear combination are given by the blending mask. Nevertheless, our study showed that transformer-based frame interpolation has the potential to increase the temporal resolution of PC imagingConclusion
We trained a transformer-based frame interpolator using a large multi-center, multi-vendor and multi-field strength dataset and demonstrate its potential in increasing the temporal resolution of 2D/4D PC MRI.Acknowledgements
No acknowledgement found.References
No reference found.Figures
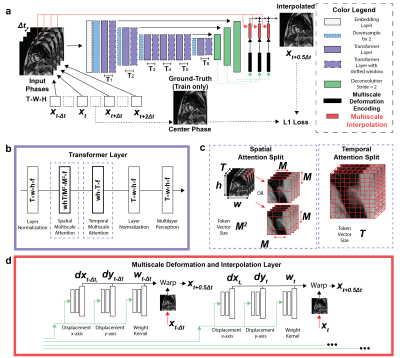
Figure 1.a)
Proposed DENT model used input W × H magnitude of cine images with W × H matrix
size of four consequent phases (i.e., T
= 4) with temporal
resolution at , , and . Output was cine image interpolated at .
For training the L1 loss was used. b)
DENT had 6 pairs of transformer layers T1-6. c) Inputs were split into windows to perform spatial (on N=w·h·T/M2
vectors of size M2×f) and temporal (N = w·h, size = T) attention.
d) Decoding convolutional layers were used to predict deformation and
scaling components used to translate and combine the 4 images onto .
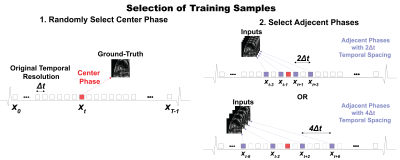
Figure 2. Selection of training samples. A sample was defined as a single-slice cine with phases
and temporal resolution; an epoch as one train loop across all
training samples. Selecting a training sample for each epoch. First, the
center-phase was randomly selected
from all available phases and was used as ground-truth. Second, adjacent input phases were chosen
either with a temporal spacing of or, equivalent to selecting phases or , accordingly. Either pattern was chosen with equal
probability.
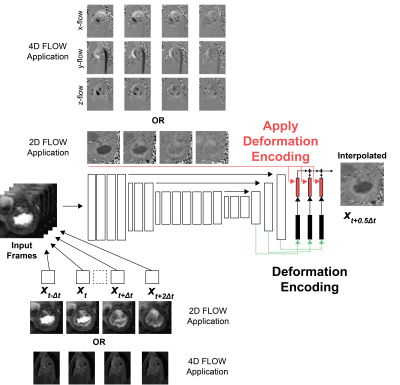
Anatomy-based encoding to interpolate 2D
or 4D flow. Flow images present with significant
salt-and-pepper noise in the image background, making direct deep learning processing
challenging. Instead, DENT enabled high-frame-rate flow using anatomy-based deformation
encodings. Specifically, DENT generated deformation and scaling components for
each anatomical input frame (shown for 2D mitral valve flow). These components
were applied to the corresponding four flow frames to synthesize a new frame.
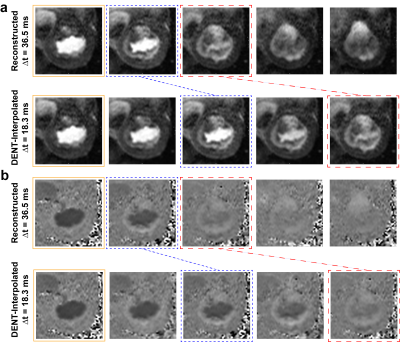
Figure 4. DENT-enabled high-frame-rate 2D flow.
Mitral valve anatomical and flow images at
temporal resolution of 36.5 ms and DENT-interpolated temporal resolution of
18.3 ms. a) First, DENT was used to
encode deformation and scaling components from anatomical images. These
encodings were used to synthesize new cardiac frames between existing ones . DENT-interpolated images enabled excellent visualization of the mitral valve
motion. b) Second, the magnitude-based
encodings were applied to flow images to synthesize new phase-contrast cine frames.
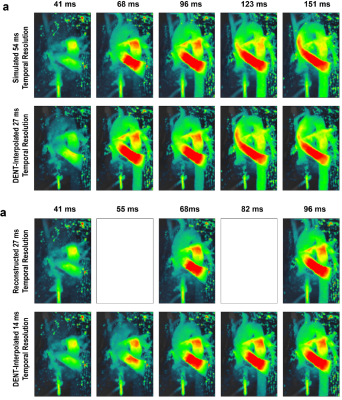
Figure 5. DENT-enabled high-frame-rate 4D
flow. a) Temporal resolution = 54 ms was simulated
by discarding every other frame (i.e., discard 41, 96 and 151 ms). DENT was
applied to remaining images to evaluate two-fold gain in temporal resolution.
DENT-interpolated images can be compared to discarded ground-truth. Note that
post-processing of 4D flow introduce variation in unaltered images. B) Alternatively, DENT was applied to
the full dataset to achieve a high-frame-rate flow of 14 ms. Empty boxes
indicate lack of prospective ground-truth.
DOI: https://doi.org/10.58530/2023/3657