3653
Improving the segmentation over time of 4D flow MRI images, using a medical image registration neural network VoxelMorph1School of Informatics Engineering, Universidad de Valparaíso, Valparaíso, Chile, 2Millennium Institute for Intelligent Healthcare Engineering, iHEALTH, Santiago, Chile, 3Department of Radiology, School of Medicine, Pontificia Universidad Católica de Chile, Santiago, Chile, 4Institute for Biological and Medical Engineering, Schools of Engineering, Medicine and Biological Sciences, Pontificia Universidad Católica de Chile, Santiago, Chile, 5Biomedical Imaging Center, Pontificia Universidad Católica de Chile, Santiago, Chile, 6School of Biomedical Engineering, Universidad de Valparaíso, Valparaíso, Chile
Synopsis
Keywords: Flow, Segmentation
The segmentation of large vessels in 4D Flow MRI remains a challenge, due to different problems such as random acquisition noise, low spatial and temporal resolution, velocity aliasing, respiratory motion, phase offsets. For that reason the use of a single segmentation has been standardized to represent the geometry throughout all the cardiac phases. However, recent studies have proposed the use of image registration to be able to solve this problem. In this work, an algorithm based on a medical image registration neural network is proposed to improve the segmentation over time for the cardiac phases of the systole period.Introduction
4D flow MRI acquisition technique allows non-invasive evaluation of blood flow velocities within a volume in the three orthogonal directions1. However, to analyze these acquisitions, the segmentation is required and remains a challenge, due to different problems such as random acquisition noise, low spatial and temporal resolution, velocity aliasing, respiratory motion, phase offsets1. For that reason the use of a single segmentation has been standardized to represent the geometry throughout all the cardiac phases. However, recent studies have proposed the use of image registration to be able to solve this problem2. In this work we adapt a 4D flow MRI processing algorithm2 using a medical image registration neural network VoxelMorph3, to improve the segmentation over time of 4D flow MRI images.Methods
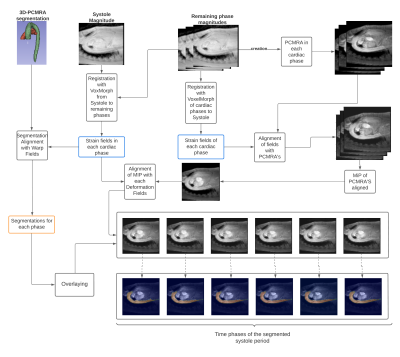
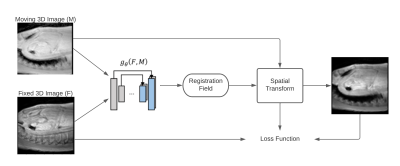
We processed 4D flow MRI data set from SIEMENS and Philips MRI scans of 32 healthy volunteers (25 men, 30.40 ± 6.23 years of age). The dataset was split into a training set belonging to 60% of the data to train the proposed neural network. This network make used of the magnitude image to perform the registration, taking as reference the magnitude image at peak de systole. The remaining 40% of the set was used to validate the variation of the segmentations in the cardiac phases. 3D phase contrast angiography (IPCMRA) was used to perform 3D segmentations of the thoracic aorta and pulmonary artery, using a 4D Flow MRI toolbox5. The training set was augmented by translations, rotations and displacements, of the magnitude images, obtained an increment of 1216 magnitude images. The summary of the registration process is shown in Figure 1.Once one segmentation was generated over the IPCMRA. We perform the time variation of that segmentation, using a similar approach proposed by Bustamante et2 .The peak systolic cardiac phase was selected as a reference, and the registration technique was modified for a medical image registration neural network technique called VoxelMorph3. The model consist in a CNN neural network with a spatial transformation layer, used to register one image from another. This allows to obtain a deformation fields from the registration step by evaluating in the learned parameters function. The loss function as $$$g_θ (F,M)=ϕ$$$ using a convolutional neural network with architecture similar to Unet6. Where $$$F$$$, $$$M$$$ are two volumes, a fixed and a moved volume, defined over a space 3-D. Where $$$ϕ$$$ is a register field and $$$θ$$$ are learnable parameters of $$$g$$$. The optimization function is defined as;
$$ϕ ̂= argmin_ϕ L(F,M,ϕ)$$
where the loss function is defined as;
$$L(F,M,ϕ)=Lsim(F,M(ϕ))+λLsmooth(ϕ)$$
$$$M(ϕ)$$$ is $$$M$$$ deformed by $$$ϕ$$$, the function $$$Lsim(·,·)$$$ measures the image similarity between $$$M(ϕ)$$$ and $$$F$$$, $$$Lsmooth(·)$$$ imposes regularization on $$$ϕ$$$, and $$$λ$$$ is the regularization parameter. The architecture of VoxelMorph is shown in Figure 2.
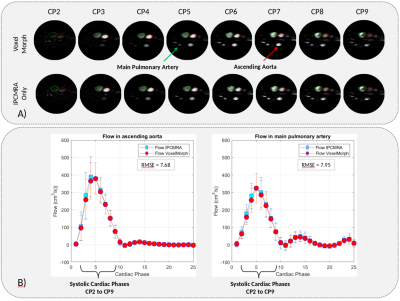
To test our algorithm, we use the segmentation of the thoracic aorta and pulmonary artery to align them in each time frame of the cardiac cycle through the deformation fields delivered by VoxelMorph using the guidelines proposed by Bustamante, et al. Obtaining the segmentations of the thoracic aorta and pulmonary artery at each time frame of the cardiac cycle. Finally, from the segmentations generated, we analyze the flow of the ascending aorta and the main pulmonary artery, from a transverse plane, using only one segmentation generated with the IPCMRA4, and the segmentations generated by VoxelMorph, the root mean square error between both curves was also calculated.
Results
We have obtained a loss during VoxelMorph training of 0.063, segmenting all cardiac phases (Figure 3). In addition we have compared the blood flow in the ascending aorta, and the main pulmonary artery (Figure 4), using the VoxelMorph segmentations and the IPCMRA segmentation, both curves are very close to each other, obtaining a mean square error of ± 7.64 in the aorta and ± 7.95 in the main pulmonary artery.Discussion
We have obtained a significant improvement in the visualization of the main anatomical regions of the cardiovascular system throughout the cardiac cycle, using VoxelMorph. One main limitation, is our images were obtained without contrast medium, for that reason, we do not have images of magnitude that allow us to segment all the cardiac phases to be able to make an objective comparison between the movement of the vessel and our segmentation, we can only compare the angiographic images of cardiac phases of the systole period, because the velocity present some changes in those cardiac phases.Our work has shown a promising approach in the segmentation of the great vessels in each time frame, using only one segmentation obtained by IPCMRA. In future contribution we want to build a deep learning neural network to generate a segmentation of the great vessel, using the IPCMRA images to automate all data processing, and we want to validate our approach with acquisitions performed with contrast agent, to increase the signal in the magnitude images.
Acknowledgements
This work has been funded by ANID: Millennium Science Initiative Program ICN2021_004. Also, FONDECYT # 1181057. Sotelo J. thanks to FONDECYT de iniciación en investigación 11200481.References
1. Dyverfeldt, Petter, et al. "4D flow cardiovascular magnetic resonance consensus statement." Journal of Cardiovascular Magnetic Resonance 17.1 (2015): 1-19.
2. Bustamante, Mariana, et al. "Improving visualization of 4D flow cardiovascular magnetic resonance with four-dimensional angiographic data: generation of a 4D phase-contrast magnetic resonance CardioAngiography (4D PC-MRCA)." Journal of Cardiovascular Magnetic Resonance 19.1 (2017): 1-10.
3. BALAKRISHNAN, Guha, et al. VoxelMorph: a learning framework for deformable medical image registration. IEEE transactions on medical imaging, 2019, vol. 38, no 8, p. 1788-1800.
4. Bock J, Frydrychowicz A, Stalder AF, Bley TA, Burkhardt H, Hennig J, Markl M. 4D phase contrast MRI at 3 T: effect of standard and blood-pool contrast agents on SNR, PC-MRA, and blood flow visualization. Magn Reson Med. 2010 Feb;63(2):330-8. doi: 10.1002/mrm.22199. PMID: 20024953.
5. Sotelo J, Mura J, Hurtado DE, Uribe S. A novel Matlab Toolbox for processing 4D flow MRI data. Proc. Intl. Soc. Mag. Reson. Med. 27 (2019). (https://github.com/JulioSoteloParraguez/4D-Flow-Matlab-Toolbox)
6. Ronneberger, Olaf, Philipp Fischer, and Thomas Brox. "U-net: Convolutional networks for biomedical image segmentation." International Conference on Medical image computing and computer-assisted intervention. Springer, Cham, 2015.
Figures