3633
3D Multichannel Convolutional Neural Network for Differentiating Benign and Malignant Solid Renal Masses with/without Bias Field Correction1Department of Radiology, University of Wisconsin Madison, Madison, WI, United States, 2Department of Biomedical Engineering, University of Wisconsin Madison, Madison, WI, United States, 3Department of Medical Physics, University of Wisconsin Madison, Madison, WI, United States
Synopsis
Keywords: Kidney, Machine Learning/Artificial Intelligence, Bias Field Correction
A deep learning model was developed for the differentiation of benign and malignant solid renal masses. A representative and balanced dataset was used, and model performance was evaluated with and without bias field correction (BFC). The model input contained multiple channels for various MRI-based tissue contrast weightings. The model achieved an accuracy and AUC of 74% and 71% with BFC and 80% and 59% without BFC. This work showed that a convolutional neural network can be trained to differentiate benign from malignant renal masses with a high degree of accuracy and that BFC improves AUC.Introduction
Solid renal masses include malignant entities, such as renal cell carcinoma (RCC) subtypes, as well as benign lesions, such as oncocytomas and angiomyolipomas (AMLs). While MRI-based techniques for distinguishing these pathologic entities have been developed [1], such techniques have not seen widespread adoption. Renal masses are often biopsied for further characterization or alternatively surgically resected without prior biopsy. Unfortunately, about 30% of resected renal masses are benign [2]. A non-invasive diagnostic tool to aid in tumor classification would be useful clinically.Artificial intelligence-based techniques for evaluating renal masses have been explored in recent years [3]. However, many studies employing these techniques have limited data sets lacking a representative cross-section of the pathology typically seen clinically or have developed models that fail to predict pathology in such a way that would mitigate the need for biopsy or surgery. The purpose of this study was to develop a 3D multichannel convolutional neural network (CNN) to determine whether a tumor is benign or malignant. Previous work in our lab has led to the curation of a balanced and representative dataset of MRI scans of the five most common solid renal mass types (oncocytomas, AMLs, and clear cell, chromophobe, and papillary RCC).
Methods
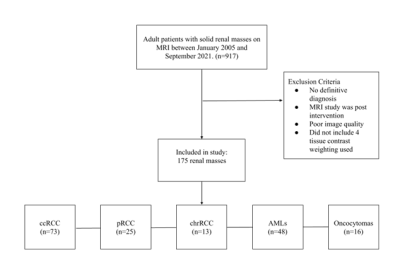
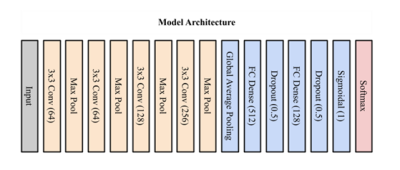
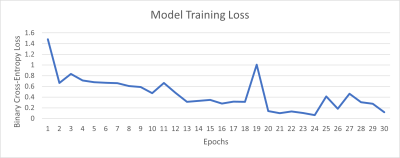
This was a retrospective study approved by the Institutional Review Board. An MRI dataset of 175 solid renal masses was curated (Figure 1). This cohort included 175 pathologically confirmed renal masses with an available pre-intervention abdominal MRI. The cohort consisted of 111 malignant tumors (73 clear cell RCC (ccRCC), 25 papillary RCC (pRCC), 13 chromophobe RCC (chrRCC)) and 64 benign tumors (48 AMLs, 16 oncocytomas). Four tissue contrast weightings were used, including T2-weighted images as well as post-contrast T1-weighted images in the arterial, 30-second delayed, and 4-minute delayed phases. Each scan underwent N4ITK bias field normalization [4] and model results using the normalized image were compared with results from the original image. Masses were segmented with a bounding box independently for each of the four tissue contrast weightings. Preprocessing of the data included cropping using the tumor segmentation followed by zero-padding for uniform input size. The data were divided 80/20 into training and testing sets. All four contrast weightings were then used to create a multichannel stack of the tumor information (Width x Height x Slice x Tissue Contrast Weighting). Training of the model occurred using both the N4ITK normalized scans and the original scans as input to determine if the N4ITK bias field normalization improved model metrics. Five-fold cross-validation was employed on the training set for both input types. The model architecture consisted of four convolutional layers with max pooling and ReLU activation followed by three dense fully connected layers (Figure 2), as previously described [3]. Training of the model occurred over 30 epochs for the multichannel approach using both input image types. The goal of the training was to minimize the binary cross-entropy loss of the model (Figure 3). The model parameters that achieved the highest validation accuracy for each fold were then selected for testing. Summative metrics included accuracy, sensitivity, specificity, positive predictive value (PPV), negative predictive value (NPV), F1 score, Matthew’s correlation coefficient (MCC), Youden’s J statistic (J), and area under the curve (AUC).Results
The best performing deep learning model used N4ITK bias field normalization on the input data, yielding an AUC = 0.71, accuracy = 0.74, sensitivity = 1.00, specificity = 0.31, PPV = 0.71, NPV = 1.00, F1 score = 0.83, MCC = 0.47, and J = 0.31. The AUC was lower when the model was trained without bias field normalization, yielding an AUC of 0.59, but an overall accuracy of 0.80 (Table 1).Discussion
The results of the model using a multichannel input were promising, achieving similar results to those found in the current literature while using a strong and representative dataset as encountered in typical clinical settings. Notably, the NPV was high, which indicates that the model successfully identifies benign masses as benign, with the potential to mitigate the need for biopsy or surgical resection in these masses. Bias field correction provides an improved AUC, which is a better metric than accuracy in the setting of class imbalances. Note that bias field correction was performed on downsampled images, which may affect results. Future work includes further training with full resolution images and bias correction.Discussion
A CNN model with bias field correction trained from MRI data can differentiate benign from malignant solid renal masses with a high degree of accuracy. Performance is improved with a multichannel approach that utilizes multiple MR tissue contrast weightings.Acknowledgements
No acknowledgement found.References
[1] I. Pedrosa and J. A. Cadeddu, "How We Do It: Managing the Indeterminate Renal Masswith the MRI Clear Cell Likelihood Score," (in eng), Radiology, vol. 302, no. 2, pp. 256-269, Feb 2022, doi: 10.1148/radiol.210034.
[2] J. H. Kim, S. Li, Y. Khandwala, K. J. Chung, H. K. Park, and B. I. Chung, “Association of Prevalence of Benign Pathologic Findings after Partial Nephrectomy with Preoperative Imaging Patterns in the United States from 2007 to 2014,” JAMA Surgery, vol. 154, no. 3. 2019, doi: 10.1001/jamasurg.2018.4602.
[3] F. Zabihollahy, N. Schieda, S. Krishna, and E. Ukwatta, "Automated classification of solid renal masses on contrast-enhanced computed tomography images using convolutional neural network with decision fusion," European Radiology, vol. 30, no. 9, pp. 5183-5190, 2020-09-01 2020, doi: 10.1007/s00330-020-06787-9.
Figures