3626
Transfer learning of renal cortex segmentation from CT to MRI: facilitated with automatic labeling1Vascular and Physiologic Imaging Research (VPIR) Lab, School of Biomedical Engineering, ShanghaiTech University, Shanghai, China, 2Central Research Institute,UIH Group, Shanghai, China, 3School of Biomedical Engineering, ShanghaiTech University, Shanghai, China, 4Shanghai Clinical Research and Trial Center, Shanghai, China
Synopsis
Keywords: Kidney, Segmentation
Segmentation of renal cortex in MR images is important but challenging. In this study, we proposed to pre-train a ResUNet model with CT images and to use an automatic method for labeling renal cortex for the training data. Such method with transfer learning and automatic labeling performed well in segmenting renal cortex in MR images, with a DICE similarity of 0.85 and volume error of 14%±5%. The proposed method would make labeling of renal cortex for training dataset much more efficiently, and we further confirm the power of transfer learning technique in segmenting renal MR images.Introduction
Accurate assessment of the various aspects of renal cortical physiology is critical, as renal cortex is where glomerular filtration is performed. For example, renal cortical perfusion is highly correlated with glomerular filtration and thus can be a good indicator of renal function1,2. Magnetic resonance imaging (MRI) has been widely adopted for renal imaging, due to its superb capability of soft-tissue imaging and multiple functional imaging methods. However, a reliable method for segmenting renal cortex in MRI images (e.g. the widely used T1-weighted images) is still missing due to a few reasons. First, renal cortex has rather a complicated shape, including both the typical cortical region and renal columns that separate the different pyramids. Second, boundary between renal cortex and medulla in human is not clear due to the distribution of different types of nephrons at different radial depths. In patients with renal dysfunction, cortical-medullary contrast can be further diminished3. For these reasons, neither manual method nor conventional segmentation method is able to segment renal cortex reliably and efficiently. A previous study demonstrated the power of transfer learning in segmenting whole kidneys in MR images4, in which a set of CT images were used to pre-train a ResUNet model. However, such method is difficult to be applied for segmenting renal cortex, because of missing labels of renal cortex for the large dataset. In this study, we proposed a simple method for automatic labeling of renal cortex, and applied the ResUNet transfer learning approach to segment renal cortex in MR T1-weighted images.Methods
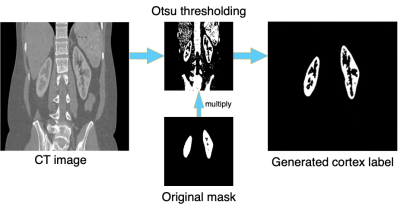
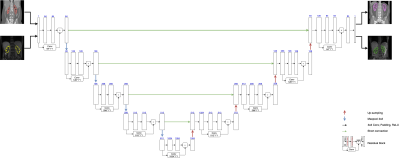
The proposed method starts with automatic generation of renal-cortex labels in CT images. Otsu thresholding5 was applied to renal CT image to get a preliminary segmentation. As the segmented mask includes both renal cortex and other regions with similar or higher intensity, we proceed to multiply it with the mask of the whole kidney. The procedure is demonstrated in Fig. 1. With this labeling technique to get renal-cortex mask for the training dataset, we trained a Deep-Residual UNet (ResUNet)6,7 (Fig. 2) to segment renal cortex. This model combines the strengths of both residual optimization and UNet8. For pre-training of this ResUNet model, we used 4000 CT images (200 subjects, each with 20 slices) with our auto-labeled cortex, and then transferred it to 60 T1-weighted MR images (6 subjects, each with 10 slices). The MR data were acquired with a 3T scanner (uMR890, United Imaging Healthcare, Shanghai).The model training on a PC workstation was equipped with a NVIDIA GeForce RTX 3070 GPU and in an environment with Ubuntu 20.04.3 LTS and Cuda 11.2. The pre-training step used 4000 CT images, with a batch size of 1 and Adam optimizer. In the fine-tuning step, we started the optimization from the pre-trained parameter values, set the learning rate to 1e-4, and trained the model with the provided 60 MRI images.
To evaluate the performance of the trained model, we used it to segment an independent set of 10 T1-weighted images and compared it against manual segmentation by an experienced human operator. For comparison, we also applied the pre-trained model (without transfer learning) and the model trained with only the 60 MRI images in segmenting the same testing dataset. DICE was used to indicate segmentation accuracy.
Results
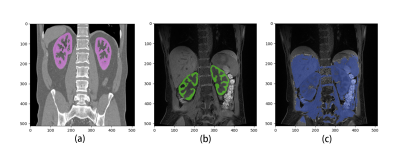
Pre-training of the ResUNet model with CT auto-labeled data was completed with a DICE accuracy of 0.93, and transfer learning to MRI images had an accuracy of 0.86. When compared to the manual segmentation across the 10 T1-weighted MR images, the proposed method had a segmentation error of 14%±5%. Fig. 3 shows representative examples of segmentation by the proposed method and by direct training (just 60 MRI images without pre-training with CT images). Without pre-training using CT images, the mask obtained with direct training mistakenly included not only the entire kidney regions but also multiple lower-abdominal organs (Fig. 3c).Discussion
While transfer learning from a large number of CT images solves the problem of small training dataset for kidney segmentation in MR images, it is still time-consuming to manually delineate the complicated mask for renal cortex for the large number of CT training datasets. Our proposed method of automatic labeling solved the problem. The model pre-trained with such labels and then fine-tuned by a small number of MR images was capable of segmenting renal cortex in MR images with high accuracy. It should be noted that the reference masks were obtained by manual segmentation, suggesting the validity of the automatic labels. The good performance of the method should also be attributed to the transfer learning of ResUNet, which managed to learn and retain the features in CT images and then to incorporate separate MR features into the final model. The approach was shown to work for segmentation of whole kidneys, and in this study works similarly well for segmenting renal cortex. The method accuracy can be further improved by eliminating the bias between labeling operators and by applying automatic labeling methods for MR training images as well.Conclusion
In conclusion, the proposed ResUNet model pre-trained with CT dataset and automatic labels of renal cortex are capable of segmenting renal cortex in MR images with relatively high accuracy.Acknowledgements
No acknowledgement found.References
1. Zollner FG, Kocinski M, Hansen L, et al. Kidney Segmentation in Renal Magnetic Resonance Imaging - Current Status and Prospects. IEEE Access. 2021;9:71577-71605. doi:10.1109/ACCESS.2021.3078430
2. Chen X, Summers RM, Cho M, Bagci U, Yao J. An Automatic Method for Renal Cortex Segmentation on CT Images. Acad Radiol. 2012;19(5):562-570. doi:10.1016/j.acra.2012.01.005
3. Lee VS, Kaur M, Bokacheva L, et al. What causes diminished corticomedullary differentiation in renal insufficiency? J Magn Reson Imaging. 2007;25(4):790-795. doi:10.1002/jmri.20878
4. Ni C, Wang Z, Lu M, Zhang JL, et al. Kidney segmentation in MR images using CT-trained ResUNet and transfer learning. Joint Annual Meeting ISMRM-ESMRMB & ISMRT 31st Annual Meeting. 2022;3697. https://archive.ismrm.org/2022/3697.html
5. Otsu N. A Tlreshold Selection Method from Gray-Level Histograms. :5.
6. Kittipongdaja P, Siriborvornratanakul T. Automatic kidney segmentation using 2.5D ResUNet and 2.5D DenseUNet for malignant potential analysis in complex renal cyst based on CT images. EURASIP J Image Video Process. 2022;2022(1):5. doi:10.1186/s13640-022-00581-x
7. Jha D, Smedsrud PH, Riegler MA, et al. ResUNet++: An Advanced Architecture for Medical Image Segmentation. ArXiv191107067 Cs Eess. Published online November 16, 2019. Accessed March 15, 2022. http://arxiv.org/abs/1911.07067
8. Couteaux V, Si-Mohamed S, Renard-Penna R, et al. Kidney cortex segmentation in 2D CT with U-Nets ensemble aggregation. Diagn Interv Imaging. 2019;100(4):211-217. doi:10.1016/j.diii.2019.03.001
Figures