3625
Size Matters: Automated Quantification of Acute Changes in Kidney Size with Deep Dilated U-Net Segmentation of Dynamic Parametric MRI1Berlin Ultrahigh Field Facility (B.U.F.F.), Max Delbrück Center for Molecular Medicine, Berlin, Germany, 2Digital Health - Machine Learning Research Group, Hasso Plattner Institute for Digital Engineering, University of Potsdam, Potsdam, Germany, 3Institute of Translational Physiology, Charité - Universitätsmedizin, Berlin, Germany, 4Hasso Plattner Institute for Digital Health, Icahn School of Medicine at Mount Sinai, New York City, NY, United States, 5Experimental and Clinical Research Center, a joint cooperation between the Charité Medical Faculty and the Max Delbrück Center for Molecular Medicine, Berlin, Germany
Synopsis
Keywords: Kidney, Preclinical
Deep learning algorithms enable fast kidney segmentation, which is crucial to establish renal size as a (pre)clinical biomarker for renal diseases. Tackling this challenge, a novel deep dilated U-Net (DDU-Net) was trained, validated, and tested on preclinical ground truth data, benchmarked on simulated data against an analytical model, and applied to longitudinal in vivo MRI scans acquired in rats, with pathophysiological interventions mimicking clinically realistic scenarios. Our DDU-Net reached a Dice score of 0.98 on the ground truth, outperformed the analytical approach, and facilitated rapid detection of acute changes in kidney size upon acute pathophysiological interventions.
Introduction
Kidney diseases are a major health issue and an increasing body of literature outlines the potential of non-invasive imaging in evaluating renal size as a clinical parameter for diagnosis, treatment, and prognosis of renal disease1-6. In vivo renal size assessment requires segmentation of the kidney. Manual segmentation is time-consuming and prone to observer bias. This constraint poses a major impediment for dynamic and longitudinal clinical and experimental studies, and severely limits the potential for renal size assessment in translational research. Machine learning (ML) approaches enable automated image segmentation7-9. The feasibility and reliability of dynamic MRI-based kidney size monitoring using ML in acute pathophysiological scenarios has not been investigated yet. Recognizing this, we examined the feasibility and reliability of MR-based kidney size (KS) assessment in rats using a novel deep dilated U-Net (DDU-Net) to facilitate quantification of acute changes in kidney size (DKS) in experimental setups mimicking realistic clinical scenarios.Methods
T2-weighted 2D images of the central coronal slice of the left kidney obtained for 52 healthy unmanipulated rats were included in this study10. A consensus reading of manual segmentation was established on baseline T2-maps of 43 rats (three maps each) as the ground truth (GT) on which the DDU-Net was trained and validated. For 29 of the 43 rats, the baseline T2-maps were additionally used for testing. The 29 animals were split randomly into three testing subsets and for each a 4-fold cross-validation was performed resulting in three ensembles of four DDU-Nets. Those 29 and the remaining 52-43=9 rats underwent six different clinically relevant interventions: occlusion of the suprarenal aorta, occlusion of the renal vein, simultaneous occlusion of both vessels, bolus injection of furosemide, short-term hypoxemia, and bolus injection of an X-ray contrast medium (CM)10-12.Compared to the original U-Net, our DDU-Net includes three extra convolutional layers with increasing dilations in each level of the encoder and decoder to extend the receptive field8, 9. Furthermore, we added intra-level skip-connections and replaced the max-pooling layers with downsampling 2x2 convolutions on the whole-level output. The final output segmentation was obtained by averaging all outputs of the DDU-Net ensemble.
The DDU-Net was evaluated against an automated bean shape model (ABSM)10 using simulated data from 400 scenarios with four contrast reduction factors (CRF) from 0.25 to 1, 10 signal-to-noise ratios (SNR) from 2.5 to 50, and 10 different ΔKS from -5% to 5%. Finally, our model was applied to six clinically relevant interventions and the ΔKS upon the respective intervention were determined and statistically analyzed.
Results
The mean KS predicted by the DDU-Net and the GT were the same with 209±1 mm2 (mean ± SEM). We determined the mean coefficient of determination (R2) = 0.94, an averaged mean absolute percentage error (MAPE) = 1%, a Dice score of 0.98, and a median intra-subject variability of 0.3% of the DDU-Net compared to 0.8% of the GT. Linear regression was performed between the predicted KS and the GT for each of the three baseline maps (Fig.1A-C) and the distribution of relative residuals between both methods determined (Fig.1D).In 247 of the 400 scenarios, the DDU-Net outperformed the ABSM (1-sided Binomial p < 10-5) in terms of MAPEs (Fig.2). For SNR > 2.5 and CRF > 0.25, in 236 out of 270 our DDU-Net outperformed the ABSM (p < 10-37) in terms of MAPEs (average of 1.2% compared to 1.6% of the ABSM), while quantifying KS more than 2 orders of magnitude faster.
For the interventions, the DDU-Net detected ΔKS = -8±1% upon occlusion of the suprarenal aorta (Fig.3A-B), ΔKS = 4±1% upon occlusion of the renal vein (Fig.3C-D), ΔKS = -2±1% upon release of the simultaneous occlusion of both vessels (Fig.3E-F), ΔKS = 2±1% after administration of furosemide (Fig.4A-B), ΔKS = -3±1% during hypoxemia (Fig.4C-D), and ΔKS of up to 9±1% for the X-ray CM bolus (Fig.4E-F).
Discussion
Even when trained on a relatively small data set, our DDU-Net offers fast, automated kidney segmentation and quantification of absolute renal size. Our findings demonstrate that the DDU-Net outperformed the analytical ABSM. Upon clinically relevant interventions the DDU-Net enabled quantification of changes in kidney size with a high accuracy.Conclusion
With ~70 ms per renal segmentation on a CPU, the DDU-Net enables real-time assessment of renal size from T2-maps. The basic principle of our approach can be extended to segmentation of the (human) kidney in 3D MR volumes. Larger datasets from population studies like the German National Cohort offer a tremendous potential to train robust ML algorithms and establish real-time KS assessment in routine clinical practice. Leveraging renal size assessment from population studies would also support deciphering the relationship between KS as a mesoscopic marker and the molecular profiles, biochemical markers and physiological measurements, with the goal of understanding the determinants and correlates of renal disease. These efforts would promote an extension of the scope of the FDA and EMA guidelines on KS assessment which are currently constrained to the use of renal size as a biomarker for autosomal dominant polycystic kidney disease. The implications of expanding the use of this marker would benefit nephrology, radiology, physiology, and related clinical fields, and help provide a foundation for new insights into renal pathology.Acknowledgements
This work was funded in part (TN, ES, KC, TG, SW) by the German Research Foundation, Project number 394046635, CRC 1365, RENOPROTECTION (Gefoerdert durch die Deutsche Forschungsgemeinschaft, Projektnummer 394046635, SFB 1365, RENOPROTECTION). We also thank the MDC-Weizmann Helmholtz International Research School for Imaging and Data Science from the NAno to the MESo (iNAMES).References
1. Hoste, E.A.J. et al. Global epidemiology and outcomes of acute kidney injury. Nat Rev Nephrol 14, 607-625 (2018).
2. Levin, A. et al. Global kidney health 2017 and beyond: a roadmap for closing gaps in care, research, and policy. The Lancet 390, 1888-1917 (2017).
3. Luyckx, V.A., Tonelli, M. & Stanifer, J.W. The global burden of kidney disease and the sustainable development goals. Bull World Health Organ 96, 414-422D (2018).
4. Global, regional, and national burden of chronic kidney disease, 1990-2017: a systematic analysis for the Global Burden of Disease Study 2017. Lancet 395, 709-733 (2020).
5. Periquito, J.S. et al. Continuous diffusion spectrum computation for diffusion-weighted magnetic resonance imaging of the kidney tubule system. Quant Imaging Med Surg 11, 3098-3119 (2021).
6. van Duijl, T.T., Ruhaak, L.R., de Fijter, J.W. & Cobbaert, C.M. Kidney Injury Biomarkers in an Academic Hospital Setting: Where Are We Now? Clin Biochem Rev 40, 79-97 (2019).
7. Sharma, K. et al. Automatic Segmentation of Kidneys using Deep Learning for Total Kidney Volume Quantification in Autosomal Dominant Polycystic Kidney Disease. Sci Rep 7, 2049 (2017).
8. Ronneberger, O., Fischer, P. & Brox, T. in Medical Image Computing and Computer-Assisted Intervention – MICCAI 2015 234-241 (2015).
9. Wang, S. et al. Stacked dilated convolutions and asymmetric architecture for U-Net-based medical image segmentation. Comput Biol Med 148, 105891 (2022).
10. Gladytz, T. et al. Reliable kidney size determination by magnetic resonance imaging in pathophysiological settings. Acta Physiol (Oxf) 233, e13701 (2021).
11. Cantow, K. et al. Quantitative Assessment of Renal Perfusion and Oxygenation by Invasive Probes: Basic Concepts. Methods Mol Biol 2216, 89-107 (2021).
12. Cantow, K. et al. Monitoring kidney size to interpret MRI-based assessment of renal oxygenation in acute pathophysiological scenarios. Acta Physiol (Oxf), e13868 (2022).
Figures
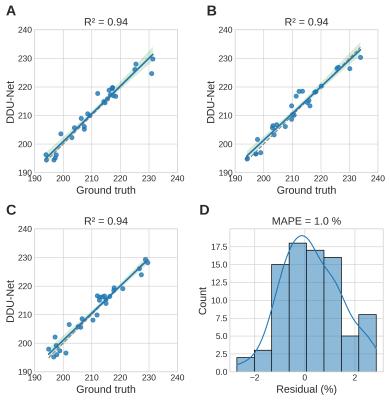
Figure 1. A-C: Linear regression plots (blue) including the 95% confidence interval (green) and the identity (gray) for the ground truth KS and predicted KS by the DDU-Net for each baseline map. D) Distribution of the residuals between ground truth and predicted KS for all three baseline maps yielding an average MAPE of 1%.
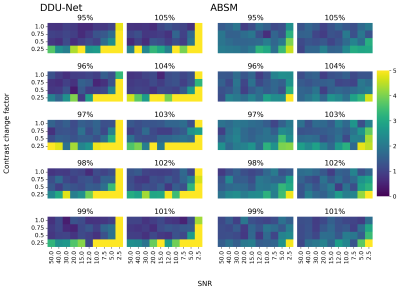
Figure 2. MAPEs achieved by the DDU-Net and the ABSM for every combination of SNR, CRF, and relative KS. For SNRs > 2.5 and CRF > 0.25, the DDU-Net (MAPE = 0-2%) outperforms the ABSM (MAPE = 1-3%).
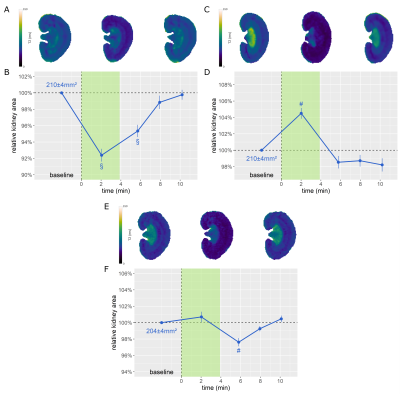
Figure 3. T2 maps acquired before, during, and after occlusion of the A) suprarenal aorta, C) renal vein, and E) both vessels. B/D/F: Relative KS over time for the respective intervention. The intervention period is highlighted in green. (Dunn’s post hoc test. With α = 0.05, a p < α/2 = 0.025 was considered to be statistically relevant. §: p < 0.001, #: p < 0.01)
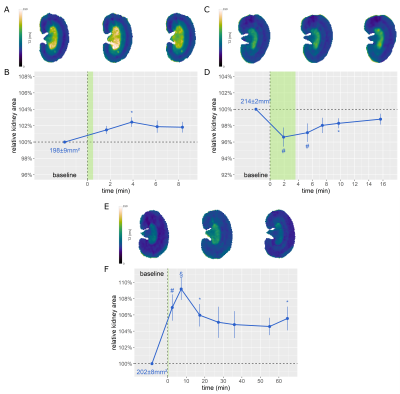
Figure 4. T2 maps acquired before, upon, and after A) administration of furosemide, C) hypoxia, and E) administration of an X-ray CM. B/D/F: Relative KS over time for the respective intervention. The intervention period is highlighted in green. (Dunn’s post hoc test. With α = 0.05, a p < α/2 = 0.025 was considered to be statistically relevant. §: p < 0.001, #: p < 0.01, *: p < 0.025)