3619
Patch-CNN provides high-fidelity directional & scalar parameter estimation from 6-directional DWI robust to pathology unseen during training
Tobias Goodwin-Allcock1, Guglielmo Genovese2,3,4, Belen Zaid3,5, Stéphane Lehericy2,3, Charlotte Rosso3,5, Ting Gong1, Robert Gray6, Parashkev Nachev6, Marco Palombo7,8, and Hui Zhang1
1Department of Computer Science and Centre for Medical Image Computing, UCL, London, United Kingdom, 2Centre de NeuroImagerie de Recherche - CENIR, Paris Brain Institute - ICM, Paris, France, 3UMR S 1127, Inserm U 1127, CNRS UMR 7225, ICM, F-75013, Sorbonne Université, Paris, France, 4Center for Magnetic Resonance Research, Department of Radiology, University of Minnesota, Minneapolis, MN, United States, 5Paris Brain Institute - ICM, Centre de NeuroImagerie de Recherche - CENIR, Paris, France, 6University College London Queen Square Institute of Neurology, London, United Kingdom, 7Cardiff University Brain Research Imaging Centre (CUBRIC), School of Psychology, Cardiff University, Cardiff, United Kingdom, 8School of Computer Science and Informatics, Cardiff University, Cardiff, United Kingdom
1Department of Computer Science and Centre for Medical Image Computing, UCL, London, United Kingdom, 2Centre de NeuroImagerie de Recherche - CENIR, Paris Brain Institute - ICM, Paris, France, 3UMR S 1127, Inserm U 1127, CNRS UMR 7225, ICM, F-75013, Sorbonne Université, Paris, France, 4Center for Magnetic Resonance Research, Department of Radiology, University of Minnesota, Minneapolis, MN, United States, 5Paris Brain Institute - ICM, Centre de NeuroImagerie de Recherche - CENIR, Paris, France, 6University College London Queen Square Institute of Neurology, London, United Kingdom, 7Cardiff University Brain Research Imaging Centre (CUBRIC), School of Psychology, Cardiff University, Cardiff, United Kingdom, 8School of Computer Science and Informatics, Cardiff University, Cardiff, United Kingdom
Synopsis
Keywords: Data Processing, Diffusion Tensor Imaging, Machine Learning
This work evaluates the clinical viability of Patch-CNN for estimating diffusion MRI (dMRI) parameters from only 6 diffusion-weighted images (DWIs). Machine learning (ML) has been proposed to improve fitting from 6-directional DWIs. However, directional measures, e.g. primary fibre orientation, have only been estimated using CNNs. CNNs have not yet been validated on pathology that is not contained within the training dataset. As pathological diversity is difficult to capture in typical applications, ML methods are clinically viable only if they can generalise to unseen pathology. We show that Patch-CNN may generalise to unseen pathology and estimate directional measures.Introduction
This work evaluates the clinical viability of Patch-CNN for estimating diffusion MRI (dMRI) parameters from only 6 diffusion-weighted images (DWIs). As parameter estimation from 6-directional DWIs is challenging with conventional fitting, machine learning (ML) has been proposed to provide an alternative. Voxel-wise neural networks1,2 are shown to improve the estimation of scalar parameters, such as fractional anisotropy (FA), and to be robust to pathology unseen during training2. However, they have not been shown to improve the estimation of directional parameters, such as the primary fibre orientation. Convolutional neural networks (CNN) are shown to improve the estimation of both directional and scalar parameters3. However, generalisation to unseen pathology is yet to be demonstrated. As pathological diversity is difficult to capture in typical applications, ML methods are clinically viable only if they can generalise to unseen pathology. This work aims to investigate whether Patch-CNN4, recently applied to the present challenge5, is generalisable to unseen pathology.Methods
To assess if Patch-CNN can generalise to unseen pathology, we train a Patch-CNN model on data excluding the pathology we used to evaluate the model performance.Network: The same Patch-CNN architecture and training parameters are used as Patch-CNN-DTI5 because this method has been shown to 1) accurately estimate scalar and directional measures from only 6 DWIs, 2) require only one training subject and 3) this architecture doesn’t require a full image as input so we do not require ‘healthy’ subjects to train, rather, we just require brain regions without lesion.
Dataset: An additional requirement for our experiment’s dataset is a high number of DWIs to create ground truth (GT) dMRI parameters, used as training targets, and to simulate an accelerated 6 directional scan, used as input to the network. Ischaemic Stroke patient data from 13 subjects acquired from the Pitié-Salpêtrière hospital and the Paris Brain Institute (ICM)6 is used for our study. This dataset provided us with a high number of DWIs consisting of 4 b=0 and 50 b=1000 images with non-colinear directions. The accelerated 6 DWI scan is imitated by finding the 6 DWI closest to the Skare7 sampling scheme. This dataset also provided us with patches that did and did-not contain lesion. These lesions are segmented by an expert radiologist. These segmentations are used to identify the subject with the smallest amount of lesion and the non-lesion data from that subject are used for training. One subject is used to validate that the network could generalise to another subject, the rest of the subjects are used for testing.
Evaluation: For benchmarking, we compare against conventional model fitting (MF) and a voxel-wise machine learning (voxel-NN) with a similar architecture to Patch-CNN except for the first layer which is reduced from a 3x3x3 kernel to a 1x1x1 fully connected layer. For qualitative evaluation, we show maps of the FA and difference maps for the scalar measures and the FA scaled colour encoded primary fibre orientation with and without lesion. For quantitative evaluation of FA, we show boxplots of the median errors for each subject over the 11 testing subjects. For quantitative evaluation of directional parameters, we show a scatter plot of the median errors for each subject containing at least 100 white-matter voxels in the lesion, total=5 subjects.
Results
Figure 1 shows the qualitative scalar measures. The most faithful estimation is provided by Patch-CNN. All of the white matter structures can be seen with greater clarity so the damage to these structures can be seen clearer than voxel-NN or MF. We see on the GT map that the lesion has resulted in decreased FA in the external capsule but normal FA in the internal capsule. Although both MF and voxel-NN have low error in this region, the noise in both images makes them less clear than Patch-CNN’s reconstruction. This result is backed up by the boxplots in Figure 3, where we see that Patch-CNN estimates with the least error over the 11 testing subjects both within the lesion and outside of it.Figure 2 shows the qualitative directional measures. Again Patch-CNN is shown to outperform both MF and voxel-NN in both the lesioned region, the internal capsule, and the non-lesion region, the corpus callosum. In both areas, the zoomed-in line image shows primary fibre orientation estimations. Estimations from Patch-CNN are superior to the other methods due to the greater coherence of the fibres and similarity to the ground truth. This result is consistent with the scatter plots in Figure 4. Here, Patch-CNN is shown to be the superior method at estimating both inside and outside the lesion as the median angular error across the testing subjects is smallest for Patch-CNN for all but one case. Interestingly the performance gap between Patch-CNN and MF decreases within the lesion. However, this is due improved performance from the MF and not decreased performance from Patch-CNN.
Discussion and Conclusion
We have shown that Patch-CNN is robust to pathology unseen during training. This is the first time accurate directional estimation from 6 DWIs in unseen pathology has been shown. Future work will extend the evaluation to include tractography, to acquire a dataset with both healthy controls and disease, and to evaluate the performance against CNNs.Acknowledgements
This work is supported by the EPSRC-funded UCL Centre for Doctoral Training in Medical Imaging (EP/L016478/1), the Department of Health’s NIHR-funded Biomedical Research Centre at UCLH and the Wellcome Trust.
Marco Palombo is supported by UKRI Future Leaders Fellowship MR/T020296/2.
References
- Golkov, V., Dosovitskiy, A., Sperl, J.I., Menzel, M.I., Czisch, M., Sämann, P., Brox, T. and Cremers, D., 2016. Q-space deep learning: twelve-fold shorter and model-free diffusion MRI scans. IEEE transactions on medical imaging, 35(5), pp.1344-1351.
- Aliotta, E., Nourzadeh, H., Sanders, J., Muller, D. and Ennis, D.B., 2019. Highly accelerated, model‐free diffusion tensor MRI reconstruction using neural networks. Medical physics, 46(4), pp.1581-1591.
- Tian, Q., Bilgic, B., Fan, Q., Liao, C., Ngamsombat, C., Hu, Y., Witzel, T., Setsompop, K., Polimeni, J.R. and Huang, S.Y., 2020. DeepDTI: High-fidelity six-direction diffusion tensor imaging using deep learning. NeuroImage, 219, p.117017.
- Li, Z., Gong, T., Lin, Z., He, H., Tong, Q., Li, C., Sun, Y., Yu, F. and Zhong, J., 2019. Fast and robust diffusion kurtosis parametric mapping using a three-dimensional convolutional neural network. IEEE Access, 7, pp.71398-71411.
- Goodwin-Allcock, T., Gong, T., Gray, R., Nachev, P. and Zhang, H., 2021, Patch-CNN-DTI: Data-efficient high-fidelity tensor recovery from 6 direction diffusion weighted imaging. Proc. Intl. Soc. Mag. Reson. Med. 30 (2021). Vancover.
- Genovese, G., Diaz-Fernandez, B., Lejeune, F.X., Ronen, I., Marjańska, M., Yahia-Cherif, L., Lehéricy, S., Branzoli, F. and Rosso, C., 2022. Longitudinal Monitoring of Microstructural Alterations in Cerebral Ischemia with in Vivo Diffusion-weighted MR Spectroscopy. Radiology, p.220430.
- Skare, S., Hedehus, M., Moseley, M.E. and Li, T.Q., 2000. Condition number as a measure of noise performance of diffusion tensor data acquisition schemes with MRI. Journal of magnetic resonance, 147(2), pp.340-352.
- Westin, C.F., Maier, S.E., Mamata, H., Nabavi, A., Jolesz, F.A. and Kikinis, R., 2002. Processing and visualization for diffusion tensor MRI. Medical image analysis, 6(2), pp.93-108.
Figures
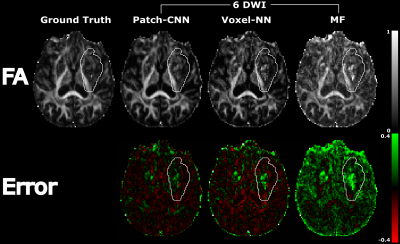
Figure 1) Maps of the FA for an individual with a stroke
lesion (outlined by the white line) covering the internal and external capsule.
We see, both inside and outside the lesion, that Patch-CNN provides the most
faithful and least noisy reconstruction of the GT image. This is affirmed by
the error plots at the bottom, where voxel-NN and MF have much greater error.
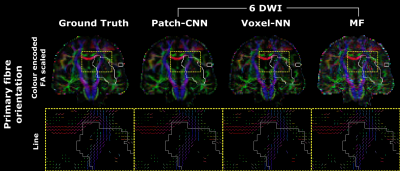
Figure 2) Top: Maps of the primary fibre orientation colour encoded
and scaled by the FA estimates. Bottom: Zoom into yellow box with primary fibre
orientation visualised as a line and coloured by direction. To remove CSF and
grey matter, only the voxels with GT FA > 0.2 were shown. In both images:
the lesion is shown by the white outline. Again, we see that Patch-CNN not only
generalises to disease unseen during training but it also provides the best
estimates. This can be seen by the coherence of the fibre orientations and how
similar they are to the ground truth.
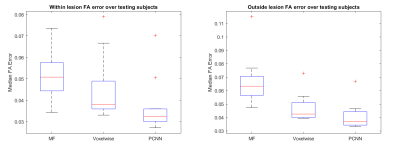
Figure 3) Boxplots of the median FA
reconstruction error of the MF, voxel-NN and Patch-CNN over 11 testing subjects.
We see that Patch-CNN far outperforms both voxel-NN and model fitting at
estimating inside and outside the lesion. This is due to the Patch-CNN having
the lowest error across all of the methods.

Figure
4) Here the primary fibre orientation error in white matter, determined by linearity8
> 0.6, is compared. This is shown in scatter of the median angular error across the 5
testing subjects, denoted by colour, whose lesion contained sufficient white matter. Patch-CNN generalises to unseen disease as the error does not decrease
within the lesion for any subject. Additionally, Patch-CNN is the most
accurate method irrespective of lesion (although marginally
better within the lesion) as Patch-CNN performs worse than MF only for the lesion data of a single subject (orange).
DOI: https://doi.org/10.58530/2023/3619