3594
High quality brain segmentation from multi-parameter mapping at Ultra-High Field MRI1Department of Physics, Norwegian University of Science and Technology (NTNU), Trondheim, Norway, 2German Center for Neurodegenerative Diseases (DZNE), Bonn, Germany, 3GIGA Cyclotron Research Centre (GIGA-CRC) in vivo imaging, University of Liège, Liège, Belgium, 4Department of Physics and Astronomy, University of Bonn, Bonn, Germany
Synopsis
Keywords: Segmentation, High-Field MRI, Multi-Parameter Mapping, Quantitative MRI, 7T
Very few segmentation techniques are optimized for 7T images. Due to the lack of approaches and the high level of inhomogeneity observed with Ultra-High Field images, neuroscientists typically limit the analysis to the upper half of the brain. A novel resolution- and contrast-agnostic technique, called SynthSeg, may remedy this limitation. A multi-parameter mapping protocol and MPRAGE images were acquired to test SynthSeg against FreeSurfer on 7T sub-millimeter brain images. Our results showed that SynthSeg can surpass FreeSurfer, especially for the cerebellum and temporal lobes. Moreover, we showed that quantitative maps can be great surrogates to high-resolution T1w images like MPRAGE.Introduction
Several algorithms exist to produce brain segmentations of MR images. Nevertheless, very few are optimized for 7T data1. Recently, convolutional neural network (CNN)-based techniques have been proposed. However, without retraining or fine-tuning, CNN-based techniques are sub-optimized. A novel segmentation technique called SynthSeg 2.02 proposes to use domain randomization3 to produce contrast- and resolution-agnostic segmentations. When high-resolution images with optimized T1 contrast (e.g., MPRAGE) are not available, SynthSeg becomes an interesting solution. The objectives of this study were twofold; 1) compare the performance of SynthSeg against FreeSurfer4 on 7T high-resolution images, and 2) evaluate the performance of SynthSeg on quantitative maps (qMaps) calculated from a 7T Multi-Parametric Mapping5 (MPM) protocol.Methods
Four healthy volunteers [2M & 2F, 22-32Y] were scanned on a 7T Siemens Magnetom Terra system (Siemens Healthineers, Erlangen, Germany) with an 8 Tx/32 Rx head coil (Nova Medical Inc., Wilmington, Delaware) in circularly polarized (CP) mode. The study was approved by the local review board and all volunteers signed a written informed consent form before the study.One MPM protocol with accelerated “skipped-CAIPI” 3D-EPI6 sampling scheme (EPI factor 4) producing images at isotropic resolution of 0.6mm was acquired. The MPM protocol included MTw, PDw, and T1w contrasts based on the combination of TRs and flip angles: 47.3 ms/7° for MTw, 33.7 ms/5° for PDw, and 33.7 ms/25° for T1w. Additionally, 4 TEs were acquired for each contrast (6.8, 13.6, 20.4 and 27.2 ms). All contrasts were acquired two times and averaged to improve SNR. Acquisition time was 16:01 min.
To correct transmit RF (B1+) inhomogeneities in the MPM images, a B1+ map was acquired from a combined slice-selective preconditioning RF pulse and turbo-FLASH readout7 with a scan time of 0:10 min.
Moreover, a (0.6mm)3 MPRAGE protocol with a TI of 1100 ms, TR of 2500 ms, FA of 5°, TE of 2.94 ms, and GRAPPA factor of 2 was acquired. Acquisition time was 8:34 min.
The MPM images and B1+ maps were given to the hMRI toolbox8 (version 0.3.0) embedded in SPM12 (http://www.fil.ion.ucl.ac.uk/spm) to calculate B1+ corrected R1, PD, MTsat, and R2* maps. Moreover, all qMaps were coregistered together in hMRI. “Synthetic MPRAGEs”9 (synMPRAGE) were created from the R1 map, and the R1 and R2* maps combined. The parameters of the synMPRAGEs were selected to maximize white and gray matter contrast10.
The MPRAGE images were bias corrected and coregistered to qMaps using a rigid registration with SPM12.
All qMaps, variations of synMPRAGEs and MPRAGE images were fed to SynthSeg, that segmented the brain into 32 subregions. No retraining or fine-tuning was performed. Additionally, MPRAGE images were also segmented with the FreeSurfer pipeline recommended for 7T datasets11. The complete pipeline is shown in Figure 1.
Results and Discussion
Example segmentations from MPRAGE images using SynthSeg and FreeSurfer-7T are shown in Figure 2. With FreeSurfer-7T, a significant section of the cerebellar cortex and bottom of brainstem were not segmented correctly, while SynthSeg was considerably more robust against the signal inhomogeneities observed in the cerebellum, brainstem, and temporal lobes. For SynthSeg, the Dice Similarity Coefficient (DSC) and 95 percentile of the Hausdorff distance (HD95) were computed between the preprocessed MPRAGE images, and ‘raw’ MPRAGE images (i.e., no bias correction) for all subregions and subjects. An average DSC of 0.98 and HD95 of 0.81 mm were calculated. Inversely, FreeSurfer-7T was unable to properly segment the ‘raw’ MPRAGE images, resulting in poor segmentation in the bottom portion of the brain. However, FreeSurfer-7T seemed slightly more sensitive to small WM-cortex boundaries compared to SynthSeg.Considering most of neuroscience research uses the 1 Tx/CP mode at 7T, our results indicate that SynthSeg will allow segmentation and quantitative analysis in the lower parts of the brain, which has been especially avoided in most studies.
As shown with Figures 3 & 4, SynthSeg was able to produce comparable segmentations for all different contrasts. The average DSC and HD95 values among subjects were computed as shown with Table 1. The R1 map and synMPRAGEs produced higher DSC and lower HD95 than the MTsat, PD and R2* maps respectively. SynthSeg was remarkably robust against the noise observed in qMaps. CSF had consistently the lowest DSC values across subjects due to the noise surrounding the brain in qMaps, making the boundary identification between CSF and skull challenging. The cerebellar cortex had systematically the largest HD95 values. Indeed, the cerebellum was noisy, resulting in suboptimal identification of the CSF-cortex and cortex-WM boundaries for qMaps. Neither the quantitative analysis nor visual assessment allowed to detect benefits of using synMPRAGEs over R1 maps.
Our results indicate that researchers working at 7T may benefit from a robust segmentation option even without high-resolution T1w images by using qMaps.
Conclusion
In this work, we found that SynthSeg can be a better option over FreeSurfer for 7T data, especially if the temporal lobes and cerebellum are of interest. Moreover, we found that qMaps can be high quality surrogates to high-resolution T1w images typically used. Ultimately, the ability of SynthSeg to segment any contrast allows us to further investigate if combining segmentations from different maps or images could produce superior segmentations.Acknowledgements
This publication is an outcome of SCAIFIELD, an EU Joint Programme - Neurodegenerative Disease Research (JPND) project (www.jpnd.eu). The project is supported through the following funding organisations under the aegis of JPND: Belgium, The Fund for Scientific Research (F.R.S.-FNRS); Germany, Federal Ministry of Education and Research (BMBF; funding codes 01ED2109A/B); Norway, The Research Council of Norway (RCN); Turkey, Scientific and Technological Research Council of Turkey, TÜBİTAK.
References
1) Svanera, M., Benini, S., Bontempi, D., & Muckli, L. (2021). CEREBRUM‐7T: Fast and Fully Volumetric Brain Segmentation of 7 Tesla MR Volumes. Human brain mapping, 42(17), 5563-5580.
2) Billot, B., Magdamo, C., Arnold, S. E., Das, S., & Iglesias, J. E. (2022). Robust segmentation of brain mri in the wild with hierarchical cnns and no retraining. In International Conference on Medical Image Computing and Computer-Assisted Intervention (pp. 538-548). Springer, Cham.
3) Tobin, J., Fong, R., Ray, A., Schneider, J., Zaremba, W., & Abbeel, P. (2017, September). Domain randomization for transferring deep neural networks from simulation to the real world. In 2017 IEEE/RSJ international conference on intelligent robots and systems (IROS) (pp. 23-30). IEEE.
4) Fischl, B. (2012). FreeSurfer. Neuroimage, 62(2), 774-781.
5) Weiskopf, N., & Helms, G. (2008). Multi-parameter mapping of the human brain at 1mm resolution in less than 20 minutes. Proceedings of 16th ISMRM, Toronto, Canada, 16, 2241.
6) Stirnberg, R., & Stöcker, T. (2021). Segmented K-space blipped-controlled aliasing in parallel imaging for high spatiotemporal resolution EPI. Magnetic resonance in medicine, 85(3), 1540–1551.
7) Chung, S., Kim, D., Breton, E., & Axel, L. (2010). Rapid B1+ mapping using a preconditioning RF pulse with TurboFLASH readout. Magnetic resonance in medicine, 64(2), 439-446.
8) Tabelow, K., Balteau, E., Ashburner, J., Callaghan, M. F., Draganski, B., Helms, G., ... & Mohammadi, S. (2019). hMRI–A toolbox for quantitative MRI in neuroscience and clinical research. Neuroimage, 194, 191-210.
9) Nöth, U., Hattingen, E., Bähr, O., Tichy, J., & Deichmann, R. (2015). Improved visibility of brain tumors in synthetic MP‐RAGE anatomies with pure T1 weighting. NMR in Biomedicine, 28(7), 818-830.
10) Tardif, C. L., Collins, D. L., & Pike, G. B. (2009). Sensitivity of voxel-based morphometry analysis to choice of imaging protocol at 3 T. Neuroimage, 44(3), 827-838.
11) Zaretskaya, N., Fischl, B., Reuter, M., Renvall, V., & Polimeni, J. R. (2018). Advantages of cortical surface reconstruction using submillimeter 7 T MEMPRAGE. Neuroimage, 165, 11-26.
Figures
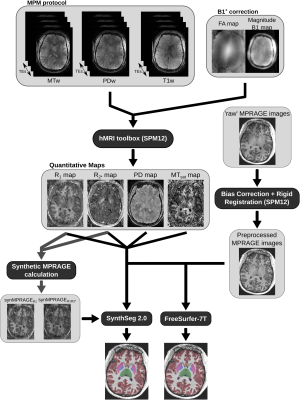
Figure 1: Schematic of the complete processing pipeline used in this study for each dataset.
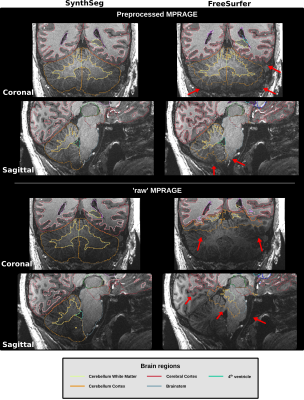
Figure 2: Visual comparison of segmentation results from SynthSeg and FreeSurfer for the same 7T (0.6mm)3 preprocessed and ‘raw’ MPRAGE images. Red arrows are showing regions where FreeSurfer was unable to segment properly.
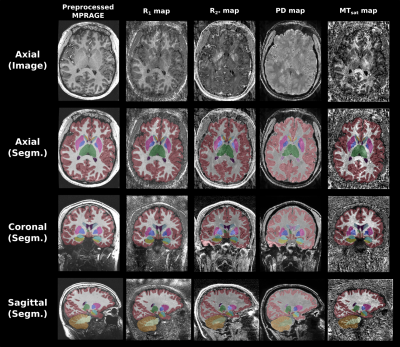
Figure 3: SynthSeg segmentations computed for the preprocessed MPRAGE images and four different qMaps tested in this work. For each input, the image/map in the axial plane is shown on the first row. The segmentation results overlapped on their corresponding input images are shown for all imaging planes on the next three rows.
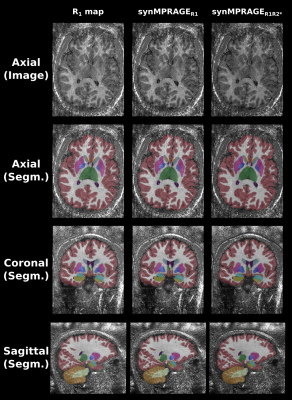
Figure 4: SynthSeg segmentations computed for the R1 map and two synMPRAGEs tested in this work. For each input, the image/map in the axial plane is shown on the first row. The segmentation results overlapped on their corresponding input images are shown for the all imaging planes on the three following rows.
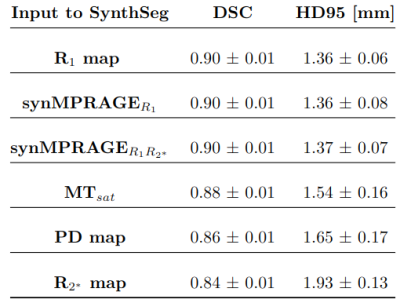
Table 1: Mean (+/- standard deviation) Dice Similarity Coefficient (DSC) and 95 percentile of the Hausdorff distance (HD95) values calculated among the four subjects for all 32 brain subregions.