3590
A new cascaded fully convolutional neural network for the simultaneous segmentation of parasagittal dural space and arachnoid granulations1Department of Neurology, Vanderbilt University Medical Center, Nashville, TN, United States, 2Department of Radiology, Vanderbilt University Medical Center, Nashville, TN, United States
Synopsis
Keywords: Segmentation, Neurofluids
The overarching goal of this work is to develop and validate novel deep learning algorithms for segmenting the peri-sinus space, including parasagittal dural (PSD) space, which has been hypothesized to harbor cerebral lymphatic channels, and intra-veinous arachnoid granulations, which has been long-hypothesized as a site a CSF egress, from standard non-contrast anatomical imaging. The new segmentation method is based on cascaded neural networks using non-contrasted 3D T2-weighted MRI; the method is method in a mixed cohort of adults with and without neurodegeneration.Introduction
The overarching goal of this work is to develop and validate novel deep learning algorithms for segmenting the parasagittal dural (PSD) space, and arachnoid granulations (AG) from non-contrast anatomical MRI. CSF flow can be characterized in terms of the traditional bulk flow pathway as well as the emerging alternative CSF efflux pathways along perivascular spaces. Classically, CSF produced in the choroid plexus (ChP) complexes within the lateral and third ventricles traverses the cerebral aqueduct en route to the 4th ventricle and to the more diffuse subarachnoid space1,2. Then, CSF traverses the subarachnoid space with resorption mediated by dural peri-sinus structures such as AGs and PSD spaces. AGs are herniations of the arachnoid membrane which penetrate the venous sinuses3. Generally, AGs have diameters ranging from millimeters to centimeters4, which are on the order of the spatial extent of typical anatomical neuroimaging voxels. Additionally, recent evidence indicates that CSF clearance can also occur in the regions surrounding the dural sinuses, the PSD5. Support for this possibility comes from intrathecal gadolinium contrast administration, whereby contrast was noted to concentrate in the subarachnoid spaces near the vertex6 followed by a progressive accumulation within the PSD. To alleviate the need of intrathecal injection of contrast agent, it has been proposed that a non-invasive MRI method may be adequate to quantify PSD in humans7. Given that intrathecal gadolinium injections are contraindicated in most research studies, such a method would have wide relevance to understanding the growing import of this structure. To investigate this possibility, a novel cascaded deep-learning method that conjointly delineate PSD and AGs from 3D T2-weighted MRI is proposed and evaluated.Methods
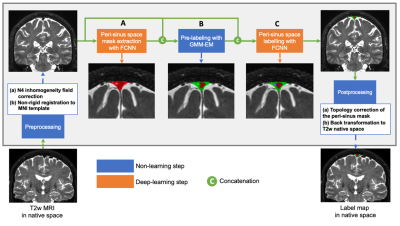
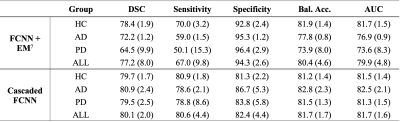
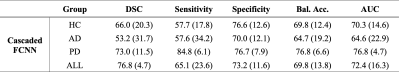
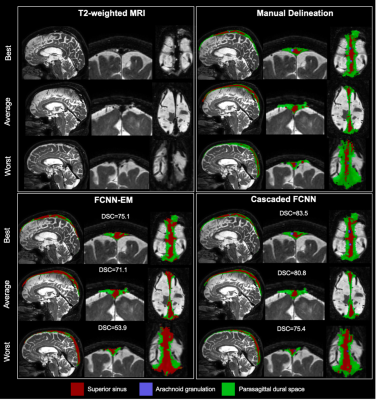
Participants comprised older adults with and without neurodegenerative pathology (Alzheimer’s disease, AD; Parkinson’s disease, PD); this heterogeneous cohort was deliberately selected to allow for increased generalizability. All participants provided informed consent and were scanned at 3T (Philips) (Table 1). Acquisition. 3D T2-weighted (T2w) imaging with spatial resolution=0.8x0.8x0.8 mm, TE=331 ms, TR=2500 ms, and sagittal acquisition. Preprocessing. T2w MRI images were corrected for inhomogeneity field8 and aligned to the MNI template using affine registration9. Algorithm. The proposed PSD segmentation method is based on a combination of extraction of a parasagittal space mask using a U-Net model10 with 3D patches as inputs (96x64x64 voxels). Pre-labelling of PSD voxels using a bi-modal gaussian mixture model, an optimal threshold computed with expectation maximization (EM) algorithm (Figure 1). A last U-Net layer infers labelling of PSD, AG using concatenation of pre-labelling, and T2w patches (cascaded-FCNN). Evaluation. PSD and AGs were manually delineated by a board certified neuroradiologist (experience=13 years), along the superior sagittal sinus extending from its most anterior aspect along the frontal calvarium to the region along the torcula posteriorly. Accuracy was evaluated using Sørensen–Dice coefficient (DSC), sensitivity, specificity, balanced accuracy, and area under the curve (AUC) in a 6-fold cross validation procedure (20 scans for training, 5 for validation, and 5 for testing), iterated 6 times. Performance were compared to prior method using a combination of deep-learning and intensity thresholding7 (FCNN+EM). Statistical differences between two methods were evaluated using paired t-tests.Results
The study comprised 29 participants different pathologies to increase generalizability (Table 1). PSD. First, automatic segmentation from our cascaded-FCNN was compared with , FCNN+EM7 (Table 2). In the healthy cohort, cascaded-FCNN and FCNN+EM provided similar performance with a DSC of 78.4 and 79.7, respectively. In the AD cohort, DSCs were 72.2 and 80.9 (p-values<0.01). In the PD cohort, DSCs were 64.5 and 79.7 (p-value<0.01). AG. The second evaluation consisted of assessing the feasibility of automatically detecting AG (Table 3). Automatic delineation of AG in the healthy cohort obtained a DSC of 66.0. In the AD cohort, DSC was 53.4; in PD cohort, DSC was 73.0.Discussions
We propose a new method that utilizes an additional deep-learning layer to accurately delineate PSD space and arachnoid granulations within the peri-sinus mask of non-invasive 3D T2w MRI scans. The accuracy of both methods was rigorously evaluated within different at-risk populations whose pathophysiology may involve the impairment of the glymphatic circuitry. This experiment shows that the novel method provides more balanced results in terms of over-/under-segmentation, but also reduces variability compared to the previous method using direct thresholding of T2w intensity. The new method is also more robust when applied to persons with neurodegeneration (i.e., AD or PD). The segmentation accuracy of the PSD space was stable across the three different groups while the experiment conducted in this work shows a significant performance increase from our previous method. We evaluated for the first time the performance for AG segmentation. This experiment showed a moderate segmentation accuracy for the three investigated populations. Finally, FLAIR sequence provides higher contrast on PSD, but fluid attenuation results in a reduced contrast in AG. Therefore, with a good contrast on both PSD and AG, T2w is well-suited to delineate these two structures.Conclusions
We propose a new method for the automatic segmentation of peri-sinus space with the accurate delineation of PSD and AG. This method provides a new tool to study a site of CSF egress, which may have relevance to CSF efflux and waste metabolism.Acknowledgements
This study has been supported in part by the National Institute of Health (NIH) through grant numbers K24-AG064114, R01-NR015079, and R01-AG062574, the Department of Defense (DoD) W81XWH-19-1-0812, and the Huntington's Disease Society of America (HDSA) HD Human Biology Project fellowship.References
1. Tumani, H., Huss, A. & Bachhuber, F. The cerebrospinal fluid and barriers – anatomic and physiologic considerations. Handb. Clin. Neurol. 146, 21–32 (2018).
2. Sakka, L., Coll, G. & Chazal, J. Anatomy and physiology of cerebrospinal fluid. Eur. Ann. Otorhinolaryngol. Head Neck Dis. 128, 309–316 (2011).
3. Jayatilaka, A. D. P. Arachnoid granulations in sheep. J. Anat. 99, 315 (1965).
4. Wolpow, E. R. & Schaumburg, H. H. Structure of the human arachnoid granulation. J. Neurosurg. 37, 724–727 (1972).
5. Absinta, M. et al. Human and nonhuman primate meninges harbor lymphatic vessels that can be visualized noninvasively by MRI. Elife 6, (2017).
6. Ringstad, G. & Eide, P. K. Cerebrospinal fluid tracer efflux to parasagittal dura in humans. Nat. Commun. 11, 1–9 (2020).
7. Hett, K. et al. Parasagittal dural space and cerebrospinal fluid (CSF) flow across the lifespan in healthy adults. Fluids Barriers CNS 1–33 (2022) doi:10.1186/s12987-022-00320-4.
8. Tustison, N. J. et al. N4ITK: Improved N3 bias correction. IEEE Trans. Med. Imaging 29, 1310–1320 (2010).
9. Avants, B. B. et al. A reproducible evaluation of ANTs similarity metric performance in brain image registration. Neuroimage 54, 2033–2044 (2011).
10. Ronneberger, O., Fischer, P. & Brox, T. U-net: Convolutional networks for biomedical image segmentation. Lect. Notes Comput. Sci. (including Subser. Lect. Notes Artif. Intell. Lect. Notes Bioinformatics) 9351, 234–241 (2015).
Figures