3586
Fully Automated Hippocampus Segmentation Pipeline using Deep Convolutional Neural Networks1Department of Neurology, Medical University Graz, Graz, Austria
Synopsis
Keywords: Segmentation, Alzheimer's Disease, Multi-Contrast, AI
Segmentation of the hippocampus on T1-weighted structural MR images is required to quantify the neurodegenerative effects in Alzheimer’s disease studies. In this work, we propose an automated artificial intelligence-based pipeline for hippocampus segmentation combined with manual ground truth (GT) data that originates from high-resolution T2-weighted MR images. Results are evaluated against the manual GT-labels and compared to the segmentation results from FreeSurfer v732. Our deep learning-based segmentation outperforms FreeSurfer in terms of accuracy and speed, while reference experiments using the T2-based GT-labels yield the best results. Thus, using T2-weighted images for ground truth generation can improve automated HC segmentation.Introduction
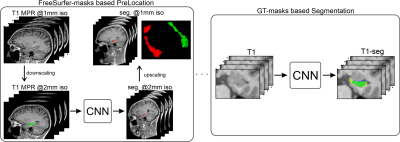
Hippocampal atrophy (tissue loss) is an important imaging biomarker in clinical trials on Alzheimer’s disease (AD). To accurately estimate the volume of the hippocampus (HC) and its progression, a robust and reliable segmentation is essential. Manual HC segmentation is considered the gold standard but is often replaced by automated software packages, such as FreeSurfer (FS). More recently, deep learning (DL) approaches have been proposed instead1,2. In most studies, HC segmentation was done on T1-weighted (T1) whole-brain MRI scans with 1mm³ isotropic resolution. However, distinct identification of the HC border on such T1 images is limited, especially near cerebrospinal fluid. A higher resolution is not feasible in clinical practice because of longer scan times and higher susceptibility for motion-induced artifacts but is crucial for ground truth (GT) annotations. We propose and evaluate the performance of a fully automated DL-based pipeline for HC segmentation on whole-brain T1 images, together with the acquisition of very high-resolution T2-weighted (T2) scans for more accurate training data (Figure 1).Methods
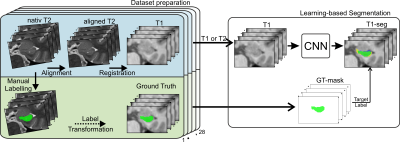
Dataset.A unique dataset was acquired to create highly accurate GT segmentations for training the neural network and establishing reference data to evaluate the performance of FreeSurfer v7.3.2. Twenty-three healthy volunteers were scanned at a Siemens Prisma 3T MR scanner to obtain corresponding pairs of high-resolution T1 and T2 images (see Figure 2).
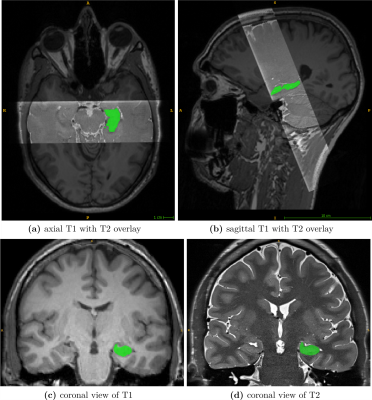
Whole-brain T1 images were acquired with a 3D magnetization-prepared rapid gradient-echo (MPRAGE) sequence (1mm³ isotropic resolution; 224x256x176mm³ field of view (FoV)). T2 scans were acquired using a 2D fast spin-echo (FSE) with hyperechoes with the oblique coronal plane perpendicular to the long axes of the hippocampi. The T2 sequence includes 40 slices with a resolution of 0.47x0.47x1mm³ (FoV, 352x512x40mm³) and covers only the hippocampal formation.
Manual GT labeling was performed on the T2 images to utilize the higher resolution and their better contrast-to-noise ratio. Therefore, the T2-based annotation protocol of Berron et al.3 was used. Overall in 18 subjects either the left or right HC was annotated, and in the remaining five subjects bilateral labeling was performed. This yields 14 manual GT-masks from each hemisphere.
To ensure agreement of the T2 scan and its GT-mask to their T1 counterpart, slice-wise alignment of the T2 slab followed by registration to the corresponding T1 scan was applied. Transformations were combined and applied at once to reduce interpolation artifacts.
Segmentation Setup.
FreeSurfer segmentations (FS-masks) were computed with the Segmentation of hippocampal subfields and nuclei of the amygdala stream4 of FS v7.3.2. To evaluate the influence of the additional T2 scan both protocols, segmentHA_T1 and segmentHA_T1T2, were applied. Moreover, FS-masks were registered back into each subject's T1-space to be comparable with the registered GT-labels.
Deep learning-based segmentation was achieved with a combination of modified U-net5 architectures. At first, we performed an initial localization of both hippocampi (see Figure 1), which is essentially a coarse segmentation trained on downsampled versions of the T1 images and the FS-masks. Second, fine segmentation of the hippocampus was achieved using our segmentation network, which consists of a stacked U-net architecture and is trained at a resolution of 0.47x0.47x1mm³ and our manual T2-based GT-labels (see Figure 3). Specifically for training the segmentation network, the dataset was preprocessed by applying slice-wise intensity normalization, flipping samples from the right to the left hemisphere. Finally, input images were cropped around the HC to a patch size of 96x64x40 pixels. Cropping was based on the center of mass of each HC and calculated from the segmentations of the initial localization.
For both networks, extensive data augmentation (translations, rotations, scaling, elastic deformations) was applied to the training data to account for anatomical variations.
Experimental Setup.
To assess the generalizability of our models, we split the preprocessed dataset (28 samples) into training (18 samples) and evaluation sets. A 3-fold cross-validation was used to account for the small dataset. All sets were balanced regarding the hemisphere.
The localization network was trained only on the T1 images. The segmentation model was trained with the following label- and image input combinations. First, T1 scans with GT-labels. Second, T2 images with GT-labels to estimate the network capabilities when T2 images are also available for segmentation. Models were trained with cross-entropy loss, ADAM optimizer, and exponential learning-rate decay for 60000 iterations with a batch size of 2.
Results
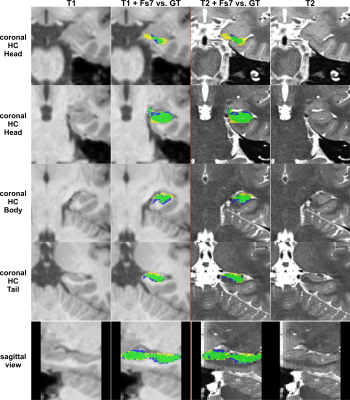
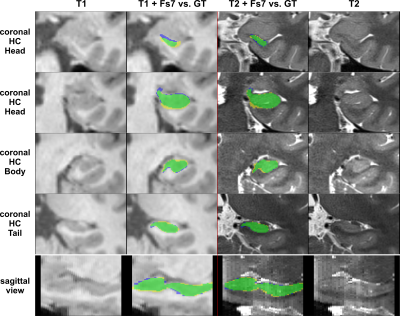
The performance of all segmentation approaches was compared via Dice similarity coefficients (DSC) against our manual GT, averaged over all cross-validation sets. HC segmentation from FreeSurfer v7.3.2 (Figure 4) achieved the lowest results, where using only T1 inputs yields 76.7±4.6% and using T1&T2 inputs results in 77.4±4.3%.Training with our GT-labels yields a DSC of 86.7±0.2% for T1 images and 92.1±0.1% for T2 images (see Figure 5).
Discussion and Conclusion
This study provides an automated pipeline for HC segmentation on T1-weighted images of large cohorts. Our approach outperforms FreeSurfer’s HC segmentation in terms of accuracy and speed (a couple of seconds vs. ~4h), regardless whether an additional T2 image for the FS pipeline was used.Moreover, using high-resolution T2-weighted scans for our network can achieve even more accurate segmentation results.
Acknowledgements
This research was supported by NVIDIA GPU hardware grants.References
1. Thyreau, B., Sato, K., Fukuda, H. & Taki, Y. Segmentation of the hippocampus by transferring algorithmic knowledge for large cohort processing. Med. Image Anal. 43, 214–228 (2018).
2. Goubran, M. et al. Hippocampal segmentation for brains with extensive atrophy using three-dimensional convolutional neural networks. Hum. Brain Mapp. 41, 291–308 (2020).
3. Berron, D. et al. A protocol for manual segmentation of medial temporal lobe subregions in 7 Tesla MRI. Neuroimage Clin 15, 466–482 (2017).
4. Iglesias, J. E. et al. A computational atlas of the hippocampal formation using ex vivo, ultra-high resolution MRI: Application to adaptive segmentation of in vivo MRI. Neuroimage 115, 117–137 (2015).
5. Ronneberger, O., Fischer, P. & Brox, T. U-Net: Convolutional Networks for Biomedical Image Segmentation. Lecture Notes in Computer Science 234–241 Preprint at https://doi.org/10.1007/978-3-319-24574-4_28 (2015).
Figures