3530
A Fully-Automatic Method to Segment Choroid Plexus in Multiple Sclerosis Using Conventional MRI Sequences
Loredana Storelli1, Elisabetta Pagani1, Martina Rubin1,2, Monica Margoni1,2, Maria Assunta Rocca1,2,3, and Massimo Filippi1,2,3,4,5
1Neuroimaging Research Unit, Division of Neuroscience, IRCCS San Raffaele Scientific Institute, Milan, Italy, 2Neurology Unit, IRCCS San Raffaele Scientific Institute, Milan, Italy, 3Vita-Salute San Raffaele University, Milan, Italy, 4Neurorehabilitation Unit, IRCCS San Raffaele Scientific Institute, Milan, Italy, 5Neurophysiology Service, IRCCS San Raffaele Scientific Institute, Milan, Italy
1Neuroimaging Research Unit, Division of Neuroscience, IRCCS San Raffaele Scientific Institute, Milan, Italy, 2Neurology Unit, IRCCS San Raffaele Scientific Institute, Milan, Italy, 3Vita-Salute San Raffaele University, Milan, Italy, 4Neurorehabilitation Unit, IRCCS San Raffaele Scientific Institute, Milan, Italy, 5Neurophysiology Service, IRCCS San Raffaele Scientific Institute, Milan, Italy
Synopsis
Keywords: Multiple Sclerosis, Neuroinflammation
Choroid plexus (CP) is a structure of the brain ventricular system that demonstrated an involvement in the inflammatory process of patients with multiple sclerosis (MS). However, its role in the pathophysiology of the disease needs to be further explored but its manual delineation on MRI is time-consuming. In this study, we developed a fully-automatic method to segment CP using 3DT1-weighted and FLAIR MRI sequences. The algorithm proved to be accurate, easy to implement and do not require an initial training phase on huge amount of data, being generalizable and a fast tool to potentially be included in a clinical setting.Introduction
Choroid plexus (CP) is a brain structure located inside the ventricular system that produces most of the cerebrospinal fluid (CSF). It has been shown to be associated with several neurological diseases including Alzheimer’s disease and stroke. There is a great interest also for the study of CP in multiple sclerosis (MS).1,2 Recent studies have shown that CP is involved in the inflammatory process and it resulted enlarged and inflamed in patients with MS.3,4 However, its role in the pathophysiology and disease evolution needs to be further explored in MS. Manual segmentation of CP is still the gold standard but it is time-consuming. Thus, the development of more accurate automatic CP segmentation methods would be very useful for the study of large cohorts. One of the freely available and well-known software for its automatic segmentation is FreeSurfer but it proved to be inaccurate.5,6 Another automatic approach has been recently proposed that, starting from FreeSurfer segmentation, clusters voxel intensities in order to separate CP from CSF and ventricular wall inside the lateral ventricles using Gaussian Mixture Model (GMM). It demonstrated good results on ADNI dataset but it starts from FreeSurfer segmentation and it was not validated on MS patients.5 A recent study based on artificial intelligence showed competitive results compared to other previously proposed deep-learning models.7 However, it was validated on a small cohort of patients and required a training phase on a significant amount of data. In this study, we proposed and validated an easily implementable fully-automatic method to segment CP using 3D T1-weighted and FLAIR brain MRI sequences. We compared our new approach to FreeSurfer and its recently proposed improvement (GMM method).Methods
We retrospectively collected baseline 3D T2-weighted Fluid attenuated inversion recovery (FLAIR) and 3D T1-weighted Magnetization prepared rapid gradient echo (MPRAGE) brain MRI for 55 MS patients (33 relapsing-remitting and 22 progressive) acquired on a 3.0 Tesla Philips Ingenia CX scanner (Philips Medical Systems). First, brain tissues were segmented on MPRAGE sequences to obtain gray matter (GM), white matter (WM) and CSF masks using FSL-SIENAX toolbox.8 The mask of the lateral ventricle in the standard MNI-152 atlas space was then registered into the subject space and used to separate peripheral to ventricular CSF. The mask of the lateral ventricle was then refined using WM and GM masks to remove hyperintense boundaries that have image intensities similar to those of the CP. The obtained ventricle mask was registered to the FLAIR space and used to extract the corresponding image intensities. A Gaussian Mixture Model (GMM) with 2 gaussian distributions was fitted to data using the iterative Expectation-Maximization algorithm. The voxels with intensities belonging to the gaussian distribution with lower values were discarded. On the survived voxels a mean filtering has been applied in order to enhance the differences between image intensities of the CP and spurious boundary voxels. A final GMM, again with two gaussian distributions, has been applied on the previously filtered mask. The proposed method has been compared with FreeSurfer and with the recently proposed GMM CP segmentation. Dice similarity coefficient (DSC), as well as volume differences and Pearson’s correlations with the manual obtained CP volumes were used to assess and compare the performance of the three automatic methods.Results
Compared to manual segmentation (Figure 1), the proposed method showed good segmentation accuracy with a mean DSC score of 0.65 (±0.08), while lower accuracy was found for Freesurfer (DSC=0.37±0.06) and GMM methods (DSC=0.38±0.08), as shown in Figure 2. The CP volumes estimated with the implemented method were very similar to the manually estimated volumes by expert physicians, with a mean percentage difference of 0.4% (±0.23%) and a slight trend to underestimate volumes (Figure 3). The mean percentage differences between CP volumes obtained by both FreeSurfer and GMM methods compared to manually extracted volumes were higher (4%±2 and 7.5%±2, respectively), showing biases of underestimation and overestimation of volumes for FreeSurfer and GMM tools respectively (Figure 3). Pearson’s correlations between automatically obtained CP volumes and the ground truth showed the highest value for our proposed method (R=0.65), and lower but comparable values for FreeSurfer (R=0.54) and GMM (R=0.56).Discussion
The fully-automatic method for CP segmentation implemented in this study proved to be highly comparable to manual segmentation performed by expert physicians, in terms of spatial overlap of the estimated structures as well as volume similarity. It showed an improvement in comparison to both FreeSurfer and the recently proposed GMM toolboxes, with a significant gain in terms of computational time (no need to run FreeSurfer segmentation). This algorithm is also easy to implement and do not require an initial training phase on huge amount of labelled data, being generalizable and a simple tool to potentially integrate in a clinical setting.Conclusion
We developed an accurate and easily implementable method for fully-automatic CP segmentation in MS patients using 3D T1-weighted and FLAIR brain MRI sequences. This algorithm could be valuable for a rapid quantification of CP volume, an additional measure of inflammation for MS patients that need further investigations.Acknowledgements
No acknowledgement found.References
1. Rodriguez-Lorenzo S, Konings J, Van Der Pol S, et al. Inflammation of the choroid plexus in progressive multiple sclerosis: accumulation of granulocytes and t cells. Acta neuropathological communications. 2020; 8(1), 1-13. 2. Vercellino M, Votta B, Condello C, et al. Involvement of the choroid plexus in multiple sclerosis autoimmune inflammation: a neuropathology study. Journal of neuroimmunology. 2008; 199(1-2), 133-141. 3. Kim H, Lim Y, Kim G, et al. Choroid plexus changes on magnetic resonance imaging in multiple sclerosis and neuromyelitis optica spectrum disorder. J Neurol Sciences. 2020; 415, 116904. 4. Ricigliano VA, Morena E, Colombi A, et al. Choroid plexus enlargment in inflammatory multiple sclerosis: 3.0-T MRI and translocator protein PET evaluation. Radiology. 2021; 204426. 5. Tadayon E, Moret B, Sprugnoli G, et al. Improving choroid plexus segmentation in the healthy and diseased brain: Relevance for Tau-PET imaging in dementia. Journal of Alzheimer’s Disease. 2020; 74(4), 1057-1068. 6. Zhao L, Feng X, Meyer C, et al. Choroid plexus segmentation using optimized 3d u-net. 2020 IEEE 17th International Symposium on Biomedical Imaging (ISBI). 2020; 381-384 IEEE. 7. Schmidt-Mengin M, Ricigliano VA, Bodini B, et al. Axial multi-layer perceptron architecture for automatic segmentation of choroid plexus in multiple sclerosis. 2022; bioRxiv. 8. Battaglini M, Jenkinson M, De Stefano N, and for the Alzheimer’s Disease Neuroimaging Initiative. SIENA-XL for improving the assessment of gray and white matter volume changes on brain MRI. Hum Brain Mapp. 2018; 39(3): 1063-1077.Figures
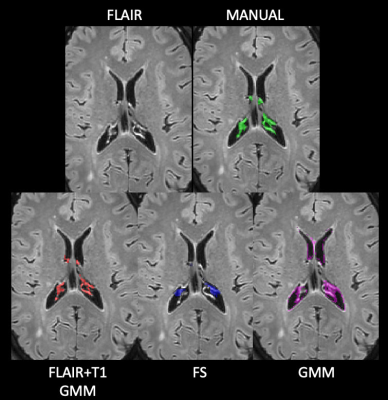
Figure 1. Examples of choroid plexus segmentations. In the
first row, an axial slice of the original FLAIR image of a patient on the left,
while on the right, the same axial slice with ground truth manual segmentation
in green. In the second row, the three automatic segmentation masks with
different colors: the proposed method in red (FLAIR+T1 GMM), FreeSurfer in blue
and the published available method in magenta (GMM). FS: FreeSurfer; GMM:
Gaussian Mixture Model.
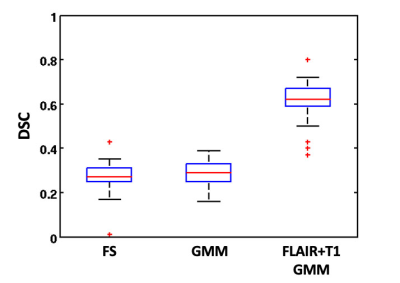
Figure 2. Boxplot of Dice similarity coefficients obtained for
the three compared methods (FS, GMM, FLAIR+T1 GMM). DSC: Dice similarity
coefficient; FS: FreeSurfer; GMM: Gaussian Mixture Model.
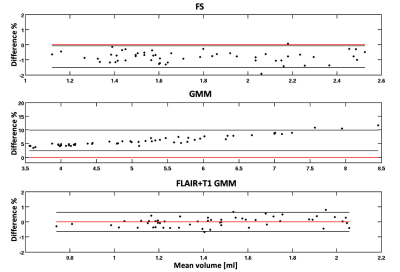
Figure 3. Bland-Altman plots of the volume percentage
differences obtained from the automatic choroid plexus segmentation methods
compared to manually obtained volumes by expert physicians. FS: FreeSurfer;
GMM: Gaussian Mixture Model.
DOI: https://doi.org/10.58530/2023/3530