3515
Diffusion tensor subspace imaging (DiTSI) delineates small fibers and gray matter microstructure invisible to single-encoding techniques1Biomedical Engineering, University of Arizona, Tucson, AZ, United States, 2Medical Imaging, University of Arizona, Tucson, AZ, United States, 3Center for Scientific Computing in Imaging, University of California, San Diego, CA, United States, 4Banner Sun Health, Sun City, AZ, United States
Synopsis
Keywords: Alzheimer's Disease, Diffusion/other diffusion imaging techniques, double diffusion encoding
Double diffusion encoding (DDE) MRI provides unique sensitization to microscale anisotropy although scalar representation of DDE data is challenging. The diffusion tensor subspace imaging (DiTSI) framework generates metrics from DDE images. In this study, post-mortem human brain tissue specimens were imaged and comparative analysis found that DiTSI but not DTI was able to provide distinct contrast in gray matter regions and at the gray/white interface. Additionally, small cranial nerve fibers of the brain stem were detectable using DiTSI, but not DTI or NODDI approaches and also showed remarkable abnormalities in Alzheimer’s disease tissue that were absent for DTI.Introduction
Measurement of specific microscale cellular features within the highly complex tissue environment of different brain structures is a major goal for diffusion MRI (dMRI) techniques to enable specific detection of pathologies. While single diffusion encoding (SDE) acquisition strategies are the most commonly used and effective for characterization of simple tissue geometries such as coherent white matter (WM), they are inherently limited in their ability to measure specific cellular features within complex tissues such as gray matter (GM). Double diffusion encoding (DDE)[1, 2] strategies in which gradients are varied in amplitude and direction relative to one another within a single acquisition period result in intrinsic sensitivity to specific microscale geometry, especially microscale anisotropy. In particular, the DDE method of diffusion tensor subspace imaging (DiTSI)[3] was developed to characterize uA in tissues for which analytical models are insufficient with two new scalar metrics, the spherical anisotropy (SA) and the radial anisotropy (RA). The objective of this study is rigorous evaluation of DiTSI metrics for their dependence on DDE encoding paradigms and for their ability to discern microscopic cellular features in GM and pathology.Methods
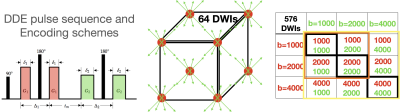
MRI microscopy was performed for healthy brain stem and temporal lobe specimens and for a brain specimen with Alzheimer’s Disease (AD) pathology and neurodegeneration. High-quality tissue specimens were obtained from the Banner Sun Health Brain and Body Donation Program[4] with short post-mortem interval and block processing. All diffusion MRI was collected with a Bruker 7T pre-clinical scanner using a 3D EPI pulse sequence with spatial resolution of 300 micron isotropic voxel dimensions. Multi-shell single diffusion encoding (SDE) weighted images with b(number directions)=1000(32), 2000(32) and 4000(56) mm2/s, and DDE weighted images were collected using a comprehensive set of sampling schemes (figure 1) in which the first and second diffusion encoding gradients were applied with b=1000, 2000 and 4000 mm2/s and every combination of b1-b2 was collected (see table in figure 1), For each set, 64 directions were acquired using cube indices to prescribe the b1 and b2 directions. The DiTSI framework was applied to calculate maps for SA and RA for comparison with DTI maps of fractional anisotropy (FA)[5] and neurite orientation distribution and dispersion imaging (NODDI)[6] maps of the orientation dispersion index (ODI).Results
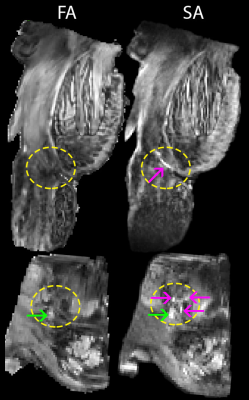
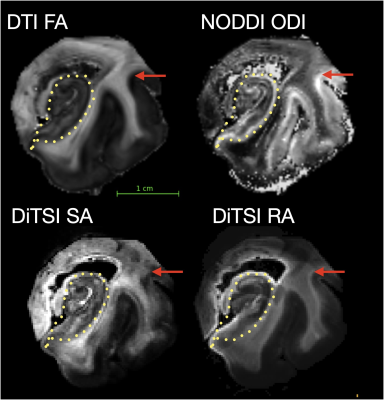
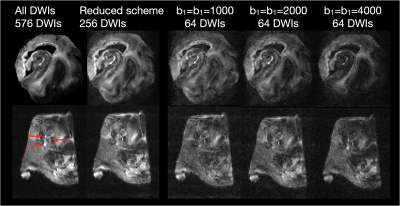
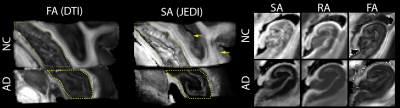
DDE acquisition and the DiTSI framework were successfully implemented to generate high-quality and high-resolution maps of SA and RA in both the brain stem and temporal lobe specimens. These showed small brain stem fibers - fascicles of cranial nerve VII - that were absent from SDE maps of any kind (figure 2). In the gray matter and at GM/WM boundaries of the temporal lobe, DiTSI metrics were distinct from DTI and NODDI metrics having greater contrast in the hippocampus and in cortical tissue (figure 3). Full DDE encoding supported maps that were rich in GM contrast and revealed small fibers, but this was also that case for a subsampled DDE scheme with less DWI volumes and a maximum b-value of 2000. For DDE with equal diffusion weighing for both encoding pulses, there was stronger GM contrast for lower b-values and greater conspicuity of small fibers for greater b-values (figure 4). Striking reduction of the DiTSI SA metric was observed in AD tissue compared with healthy control unlike for the DTI metric of FA (Figure 5).Discussion
The scalar maps of the DiTSI method were effective in detection and representation of two key features enabled by DDE acquisition and absent from SDE diffusion MRI techniques. First and most striking was the appearance of cranial nerve fascicles on SA and RA maps that were completely absent for FA or any other SDE derived map. The sensitivity of DDE strategies to small dimensional features is predicted by theory, but implementation has been challenging on a practical level and DiTSI appears to provide a set of scalar maps that accomplish this goal.The relatively high contrast of DiTSI metrics within GM structures of the brain, which have a much higher degree of microstructural complexity also suggests that this approach improves specificity to microscale anisotropy. The distinction between the ODI metric and the SA metric suggest complementary scalar metrics by these two approaches to describe microscopic structure by SA and fiber geometry population features by ODI.The DDE sampling schemes investigated spanned a broad range of b-vaules and combinations of b1-b2 weightings with small fibers most evident for high b-values and GM contrast improved for lower b-values. A reduced sampling scheme of 4 b1-b2 combinations appeared to provide similar quality and contrast to the full 9 combination scheme.Finally, SA was remarkably reduced in GM regions of a brain specimen with AD pathology and FA was not, which suggests sensitivity of the DDE acquisition and DiTSI analytic framework for the detection of microscale pathologic features.Conclusions
The DiTSI framework reveals microstructural anisotropy within complex brain tissue environments that is not apparent using single diffusion encoding techniques including cranial nerve fibers, GM microstructure and pathology in AD.Acknowledgements
All imaging was performed in the UA translational bioimaging resource (TBIR) and made possible by the NIH small instrumentation grant: S10 OD025016. This research was supported in part by the NIA/NIH grant R03 780250. All image processing was performed using the UA High Performance Computing (HPC) resources. We are grateful to the Banner Sun Health Research Institute Brain and Body Donation Program of Sun City, Arizona for the provision of human biological materials. The Brain and Body Donation Program has been supported by the National Institute of Neurological Disorders and Stroke (U24 NS072026 National Brain and Tissue Resource for Parkinson’s Disease and Related Disorders), the National Institute on Aging (P30 AG19610 and P30AG072980, Arizona Alzheimer’s Disease Center), the Arizona Department of Health Services (contract 211002, Arizona Alzheimer’s Research Center), the Arizona Biomedical Research Commission (contracts 4001, 0011, 05-901 and 1001 to the Arizona Parkinson's Disease Consortium) and the Michael J. Fox Foundation for Parkinson’s Research.References
1. Mitra, P.P., Multiple wave-vector extensions of the NMR pulsed-field-gradient spin-echo diffusion measurement. Physical Review B, 1995. 51(21): p. 15074.
2. Callaghan, P.T., NMR imaging, NMR diffraction and applications of pulsed gradient spin echoes in porous media. Magnetic resonance imaging, 1996.
3. Frank, L.R., B. Zahneisen, and V.L. Galinsky, JEDI: Joint Estimation Diffusion Imaging of macroscopic and microscopic tissue properties. Magn Reson Med, 2020. 84(2): p. 966-990.
4. Beach, T.G., et al., Arizona Study of Aging and Neurodegenerative Disorders and Brain and Body Donation Program. Neuropathology, 2015. 35(4): p. 354-89.
5. Pierpaoli, C. and P.J. Basser, Toward a quantitative assessment of diffusion anisotropy. Magnetic resonance in medicine, 1996. 36(6): p. 893-906.
6. Zhang, H., et al., NODDI: practical in vivo neurite orientation dispersion and density imaging of the human brain. NeuroImage, 2012. 61(4): p. 1000-1016.
Figures