3472
Deep Learning-Based Affine Medical Image Registration - A Review and Comparative Study on Generalizability1Computer Assisted Clinical Medicine, Medical Faculty Mannheim, Heidelberg University, Mannheim, Germany, 2Mannheim Institute for Intelligent Systems in Medicine, Medical Faculty Mannheim, Heidelberg University, Mannheim, Germany
Synopsis
Keywords: Machine Learning/Artificial Intelligence, Data Processing, Image Registration
In this research we investigated the performance of published neural networks for an affine registration of multimodal medical images and examined the networks' generalizability to new datasets. The neural networks were trained and evaluated using a synthetic multimodal dataset of three-dimensional CT and MRI volumes of the liver. We compared the Normalised Mutual Information, Dice coefficient and the Hausdorff distance across the neural networks described in the papers, using our CNN as a benchmark and the conventional affine registration method as a baseline. Seven networks improved the pre-registration Dice coefficient and are therefore able to generalise to new datasets.
Introduction
Due to the local lesion heterogeneity in patients with oligometastics disease survival rates using common therapy strategies is low. However, employing personalized approaches including minimal invasive image-guide interventions incorporating knowledge on the tumour heterogeneity significantly improves survival1,2, commonly performed using computer tomography (CT) imaging while deriving tumour heterogeneity using MRI.Therefore, registered images of CT and MRI is warranted. Architectures of several neural networks for medical image registration have been proposed recently. However, as different datasets were used to train and evaluate the networks, a fair comparison is not possible. The objective of this work is to analyse and assess the performance of several neural networks for multimodal affine image registration and to evaluate the generalisability of these networks to new datasets. Therefore, 20 different neural networks were implemented and compared using a synthetic multimodal dataset of three-dimensional CT and MR images of the liver.
Methods
We used a synthetic dataset of 112 three-dimensional CT and MR T1-weighted liver volumes. These were created using a CycleGAN3 on images of the XCAT phantom4.Each CT volume is reconstructed to a 512x512 matrix with 82-124 slices and a resolution of 1x1x2mm3. MR volumes were created with a matrix of 330x450 and 54-83 slices to match a resolution of 1x1x3mm3. This dataset contains anatomical segmentation masks for the liver.
The CT and MR images are available in two breathing states: inhaled and exhaled. The MR images are registered affinely to the CT images, inhaled state to exhaled state, and vice versa.
Networks Architectures
We implemented 20 neural networks for affine registration of medical images found on IEEE Xplore, PubMed, Web of Science and ScienceDirect with the keywords affine, registration, deep learning, artificial neural network, medical images. The architectures can be broadly grouped into different categories, depicted in figure 1. In some papers, parts of the networks' implementation were unclear, especially parameter settings were not reported making the reproducibility difficult. Assumptions were made for these parameters, and settings commonly used in affine registration network architectures, e.g. a kernel size of 3x3x3, were chosen.
Benchmark Neural Network and Baseline method
As benchmark, we used our own developed Convolutional Neural Network (CNN) (figure 2). As baseline, i.e. as state-of-the-art, we applied the Elastix affine registration algorithm implemented in the SimpleElastix Toolbox using default parameters. Mutual Information is used as objective function.
Training Setting
Training of the networks is performed with semi-supervised learning according to the VoxelMorph framework8. During training, in addition to the fixed and moving images, the segmentations of the fixed and moving images are given as input. We used the Dice loss function on the segmentations.
We performed five-fold cross-validation and trained the networks for 100 epochs with a batch size of one, stopping after five epochs with no improvement in validation loss. Adam optimiser was used with a learning rate of 1e-4.
We resized the network's inputs to 256x256x64 voxels. The images were used in half resolution due to GPU memory limitations. The resulting affine transformation was applied to the full resolution images. During preprocessing, the image intensity was normalised to the range [0,1].
Evaluation Metrics
To quantify structural differences, we applied Normalised Mutual Information (NMI) on the fixed and result (moved) image. We measured the overlap of the segmentations of the moved and fixed image using the Dice coefficient and the Hausdorff distance (HD).
Results
No neural network yielded an NMI higher than the NMI of the unregistered images (pre-registration NMI). Seven out of 20 networks improved the Dice coefficient and five networks were able to reduce the Hausdorff distance. Table 1 summarizes the results. The network of Gu et al. (2019)10 achieved the highest Dice coefficient. Our benchmark network yielded a similar Dice coefficient (figure 3). The neural network proposed by Hasenstab et al. (2019)7 failed to register the data at all. The image intensities of the moved image and its segmentation are set completely to background value.Discussion
We implemented 20 different machine learning-based image registration methods and evaluated them with respect to an affine multimodal registration of CT and MRI images of the liver. Furthermore, we implemented a framework to test and evaluate these approaches systematically.Our results show that although each network was trained on the synthetic data only few could generalize, i.e. adapt to the new data and produce good registration results compared to a conventional affine registration algorithm and an inhouse developed deep learning approach.
To allow a fair comparison, we tried to reproduce the algorithms as best as possible, however, in some papers, not all parameters for the neural networks’ implementation were described. This is detrimental to the reproducibility and assumptions were made, that may limit possible better registration results.
So far, we only investigated affine transformations applied to synthetically generated data. Future work will focus on patient data and other transformations, e.g. deformable registration.
Conclusion
Although various machine learning algorithms for affine multimodal image registration are proposed, few can generalize to new data and applications. More work should be invested in developing more general applicable image registration techniques to be able to be used in minimal-invasive image guided interventions to support precision medicine in cancer treatment.Acknowledgements
This research project is part of the Research Campus M2OLIE and funded by the German Federal Ministry of Education and Research (BMBF) within the Framework "Forschungscampus: public-private partnership for Innovations" under the funding code 13GW0388A.
This project was supported by the German Federal Ministry of Education and Research (BMBF) under the funding code 01KU2102, under the frame of ERA PerMed.
References
1. Qiu, H., Katz, A.W., Milano, M.T., 2016. Oligometastases to the liver: predicting outcomes based upon radiation sensitivity. J Thorac Dis (10):E1384-E1386. doi:10.21037/jtd.2016.10.88.
2. Ruers, T., Van Coevorden, F., Punt, C., Pierie, J., Borel-Rinkes, I., Ledermann, J., Poston, G., Bechstein, W., Lentz, M., Mauer, M., Folprecht, G., Van Cutsem, E., Ducreux, M., Nordlinger, B., 2017. Local treatment of unresectable colorectal liver metastases: Results of a randomized phase II trial. J Natl Cancer Inst 109(9):djx015. doi:10.1093/jnci/djx015.
3. Bauer, D.F., Russ, T., Waldkirch, B.I., Tönnes, C., Segars, W.P., Schad, L.R., Zöllner, F.G., Golla, A.K., 2021. Generation of annotated multi-modal ground truth datasets for abdominal medical image registration. International Journal of Computer Assisted Radiology and Surgery 16, 1277–1285. doi:10.1007/s11548-021-02372-7.
4. Segars, W., Sturgeon, G., Mendonca, S., Grimes, J., Tsui, B., 2010. 4d xcat phantom for multimodality imaging research. Medical physics 37, 4902–15. doi:10.1118/1.3480985.
5. Chen, J., Frey, E.C., He, Y., Segars, W.P., Li, Y., Du, Y., 2021a. Transmorph: Transformer for unsupervised medical image registration. doi:10.48550/ARXIV.2111.10480.
6. Mok, T.C.W., Chung, A.C.S., 2022. Affine medical image registration with coarse-to-fine vision transformer. doi:10.48550/ARXIV.2203.15216.
7. Hasenstab, K.A., Cunha, G.M., Higaki, A., Ichikawa, S., Wang, K., Delgado, T., Brunsing, R.L., Schlein, A., Bittencourt, L.K., Schwartzman, A., Fowler, K.J., Hsiao, A., Sirlin, C.B., 2019. Fully automated convolutional neural network-based affine algorithm improves liver registration and lesion co-localization on hepatobiliary phase t1-weighted mr images. Eur Radiol Exp. 3(1):43. doi:10.1186/s41747-019-0120-7.
8. Balakrishnan, G., Zhao, A., Sabuncu, M.R., Guttag, J., Dalca, A.V., 2019. VoxelMorph: A learning framework for deformable medical image registration. IEEE Transactions on Medical Imaging 38, 1788–1800. doi:10.1109/tmi.2019.2897538.
9. Hu, Y., Modat, M., Gibson, E., Li, W., Ghavami, N., Bonmati, E., Wang, E., Bandula, S., Moore, C.M., Emberton, M., Ourselin, S., Noble, J.A., Barratt, D.C., Vercauteren, T., 2018. Weakly-supervised convolutional neural networks for multimodal image registration. Medical Image Analysis 49, 1–13. doi:10.1016/j.media.2018.07.002.
10. Gu, D., Liu, G., Tian, J., Zhan, Q., 2019. Two-stage unsupervised learning method for affine and deformable medical image registration, in: 2019 IEEE International Conference on Image Processing (ICIP), pp. 1332–1336. doi:10.1109/ICIP.2019.8803794.
11. Shen, Z., Han, X., Xu, Z., Niethammer, M., 2019. Networks for joint affine and non-parametric image registration. doi:10.48550/ARXIV.1903.08811.
12. Zhao, S., Lau, T., Luo, J., Chang, E.I.C., Xu, Y., 2020. Unsupervised 3d end-to-end medical image registration with volume tweening network. IEEE Journal of Biomedical and Health Informatics 24, 1394–1404.doi:10.1109/jbhi.2019.2951024.
13. Luo, G., Chen, X., Shi, F., Peng, Y., Xiang, D., Chen, Q., Xu, X., Zhu, W., Fan, Y., 2020. Multimodal affine registration for icga and mcsl fundus images of high myopia. Biomedical Optics Express 11. doi:10.1364/BOE.393178.
14. Tang, K., Li, Z., Tian, L., Wang, L., Zhu, Y., 2020. Admir–affine and deformable medical image registration for drug-addicted brain images. IEEE Access 8, 70960–70968. doi:10.1109/ACCESS.2020.2986829.
15. Zeng, Q., Fu, Y., Tian, Z., Lei, Y., Zhang, Y., Wang, T., Mao, H., Liu, T., Curran, W., Jani, A., Patel, P., Yang, X., 2020. Label-driven mri-us registration using weakly-supervised learning for mri-guided prostate radiotherapy. Physics in Medicine & Biology 65. doi:10.1088/1361-6560/ab8cd6.
16. Chen, X., Meng, Y., Zhao, Y., Williams, R., Vallabhaneni, S.R., Zheng, Y.,2021b. Learning Unsupervised Parameter-Specific Affine Transformation for Medical Images Registration. pp. 24–34. doi:10.1007/978-3-030-87202-1 3.
17. Gao, X., Van Houtte, J., Chen, Z., Zheng, G., 2021. Deepasdm: a deep learning framework for affine and deformable image registration incorporating a statistical deformation model, in: 2021 IEEE EMBS International Conference on Biomedical and Health Informatics (BHI), pp. 1–4.doi:10.1109/BHI50953.2021.9508553.
18. Roelofs, T.J.T., 2021. Deep Learning-Based Affine and Deformable 3D Medical Image Registration. Master’s thesis. Aalto University. Espoo, Finnland.
19. Shao, W., Banh, L., Kunder, C.A., Fan, R.E., Soerensen, S.J.C., Wang, J.B., Teslovich, N.C., Madhuripan, N., Jawahar, A., Ghanouni, P., Brooks, J.D., Sonn, G.A., Rusu, M., 2020. Prosregnet: A deep learning framework for registration of mri and histopathology images of the prostate. doi:10.48550/ARXIV.2012.00991.
20. Zhu, Z., Cao, Y., Chenchen, Q., Rao, Y., Di, L., Dou, Q., Ni, D., Wang, Y., 2020. Joint affine and deformable 3d networks for brain mri registration. Medical Physics 48. doi:10.1002/mp.14674.
21. Chee, E., Wu, Z., 2018. Airnet: Self-supervised affine registration for 3d medical images using neural networks. doi:10.48550/ARXIV.1810.02583.
22. de Vos, B.D., Berendsen, F.F., Viergever, M.A., Sokooti, H., Staring, M., Išgum, I., 2019. A deep learning framework for unsupervised affine and deformable image registration. Medical Image Analysis 52, 128–143. doi:10.1016/j.media.2018.11.010.
23. de Silva, T., Chew, E.Y., Hotaling, N., Cukras, C.A., 2020. Deep-learning based multi-modal retinal image registration for longitudinal analysis of patients with age-related macular degeneration. Biomedical Optics Express 12. doi:10.1364/BOE.408573.
24. Venkata, S.P., Duffy, B.A., Datta, K., 2022. An unsupervised deep learning method for affine registration of multi-contrast brain mr images.
25. Waldkirch, B.I., 2020. Methods for three-dimensional Registration of Multimodal Abdominal Image Data. Ph.D. thesis. Ruprecht Karl University of Heidelberg.
Figures

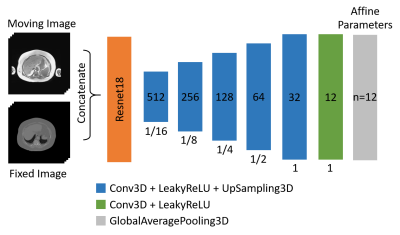
Figure 2: Architecture of the benchmark CNN for affine registration. A Resnet18 encoder and a U-Net-like decoder with one convolution per layer without skip connections. The decoder consists of six three-dimensional convolutions with a kernel size of 3x3x3, a stride of 1 and padding same. Global Average Pooling then is applied to generate the 12 parameters needed for a three-dimensional affine transformation.
Table 1: The Normalized Mutual Information (NMI), Dice coefficient and Hausdorff distance (HD) results (mean ± SD) on our synthetic dataset for the implemented networks, the benchmark (our CNN) and the baseline (SimpleElastix affine). NMI: higher value is better (maximum is 1), Dice coefficient: higher value is better (maximum is 100%), Hausdorff distance: lower value is better (minimum is 0).
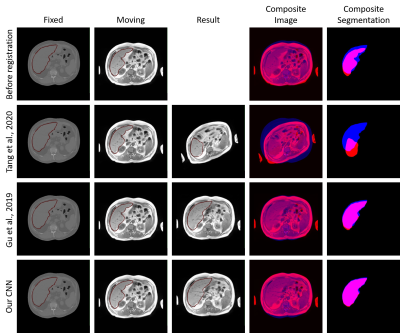
Figure 3: Example results of an affine registration of two volumes from the synthetic dataset. The images show central slices of the axial plane. The fixed images, moving images and result (moved) images are overlaid by the liver segmentations (column 1 - 3). The resulting composites for image and segmentation show the fixed data in blue and the moving/moved data in red. The second row illustrates an example of a poor registration result. Row three and four represent good registration results.