3443
Parametrized macromolecule models in the brain 1H MRS quantification: Effect of the baseline spline on quantification uncertainty and precision1Charité – Universitätsmedizin Berlin, corporate member of Freie Universität Berlin and Humboldt- Universität zu Berlin, Department of Neuroradiology, Berlin, Germany, Berlin, Germany, 2Physikalisch-Technische Bundesanstalt (PTB), Braunschweig und Berlin, Germany., Berlin, Germany, 3LGC Limited Registered, Teddington, Middlesex, United Kingdom
Synopsis
Keywords: Data Analysis, Spectroscopy, macromolecules, single-voxel 1HMRS
Parametric macromolecule (MM) models can improve the quantification of brain 1H-MRS. We quantified a publicly available repeated-acquisition dataset with LCModel and applied a parametric MM model in conjunction with four different settings for spline node distance and compared the quantification results. Coefficients of variation, average Cramér-Rao lower bound, repeatability, and reproducibility were estimated. A moderate effect of the baseline spline was shown.Introduction
MR spectroscopy (MRS) is the only non-invasive technique capable of detecting metabolite concentration changes in living tissues and thus giving an insight into diseases at the molecular level. The use of ultra-high field strengths and short echo-times allow to acquire spectra with high resolution. However, a higher resolution leads to an increased spectral complexity, and it becomes crucial to apply an approach capable of accurately fitting many components. Moreover, because in many diseases such as Alzheimer’s1 metabolite concentration differences are subtle, the determination of precision of the method is of primary importance to enable proper data interpretation. Precise quantification of brain 1H-MRS is challenging due to a background of broad resonances from macromolecules (MM). Multiple methods were proposed in the past years to consider the MM, most of them involve the use of multiple MM components parametrized with soft-constraints3–5,12. As these methods increase the degrees of freedom of the model, avoiding overfitting, and hence, overinterpretation of data, is challenging. The estimation of variance components in a repeated-acquisition dataset allows to overcome this challenge. LCModel makes use of a spline baseline, with a configurable nodes distance in ppm. Studies investigating the influence of this baseline are rare4,6–8. To our best knowledge, no study used repeated-acquisition to estimate this contribution. This study aims to examine the influence of the baseline stiffness in LCModel on the metabolite quantification. To this end, we used a repeated-acquisition dataset. To compare the impact of four different spline settings, the coefficients of variation (CVs), the Cramér-Rao lower bounds (CRLBs), as well as the reproducibility and repeatability on the estimated concentrations are calculated.Method
An open-dataset9,10 containing repeated-acquisition SPECIAL11-acquired 7T data from the posterior cingulate cortex of nine volunteers was used. Spectra were processed using a MATLAB tool including the following steps: Summation of odd and even acquisitions, weighted and phase-corrected coil-element combination, frequency correction and averaging. A parametrized-ratio MM model (PRaMM)12 of 15 MM peaks and 13 intensity ratios of high correlated MM peaks as soft-constraints is used (LCModel parameters CHSIMU and CHRATO). The water concentrations, calculated from the voxels’ tissue fractions, and the correction for tissue water relaxation within GM and WM are assigned to the LCModel parameters WCONC and ATTH2O respectively. Four different settings of the DKNTMN parameter, which fixes the distance of the baseline spline nodes were considered: 0.15 ppm (PRaMM015); 0.25 ppm (PRaMM025); 1 ppm (PRaMM1); and 99 ppm (PRaMM99). Finally, absolute concentrations were calculated, by correcting the LCModel output data for CSF fraction and metabolite relaxation times. CVs on concentrations and CRLBs, averaged across sessions, are tested for differences with the Friedman test13 and the Wilcoxon signed-rank post-hoc14 test. p-values≤0.05 were considered significant. Bonferroni correction was applied when necessary. To quantify the precision of each model, a restricted maximum likelihood (REML) estimation was conducted15, accounting for the unbalanced nested study design in which the data set was acquired. The variance components were extracted separately as standard deviations (SD) for each metabolite/postprocessing combination. For each metabolite, the models were tested pairwise for differences using the Tukey’s test. A Friedman test on the residual SDs on repeatability and reproducibility was performed to determine the significance on precision.Results and Discussion
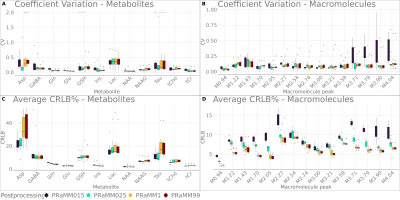
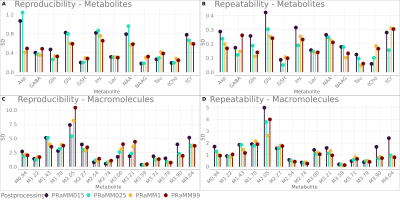
Averaged LCModel quantification results are shown in Fig.1. Friedman test showed a significant -but small in magnitude– effect of spline node distances only on MM repeatability. The analyses with a spline node distance<1ppm exhibit similar results, as do the analyses with spline node distances≥1ppm. Tukey’s test on the REML models revealed no significant differences between PRaMM99 and PRaMM1. Significant differences between PRaMM025 and PRaMM1, as well as between PRaMM025 and PRaMM99 were found for Asp, GABA, Glu, Ins, NAA, Tau, tCr, M2.27, M3.59, M3.71, M3.79, M3.90 and M4.04. Almost all estimated metabolite concentrations exhibited a significant difference, when the PRaMM015 settings were compared to both PRaMM1 and PRaMM99. Moreover, significant differences were observed between PRaMM025 and PRaMM015 for all metabolites except GABA, NAA, NAAG and all MM except M1.43 and M3.59. PRaMM015 and PRaMM025 exhibit a lower repeatability and reproducibility of metabolite concentrations (Fig3A,B) compared to PRaMM1 and PRaMM99. Moreover, the smaller spline node distances lead to significantly higher CRLBs for almost all MM signals (Fig.2D) compared to settings with higher distances, while the CRLBs for most metabolites are significantly lower. This indicates a potential overfitting resulting from too many degrees of freedom of the baseline fit7. This effect is less pronounced in PRaMM025. PRaMM1 and PRaMM99 result in similar CVs and CRLBs for almost all metabolites and MM. They appear to have a higher repeatability of the MMs compared to PRaMM015 and PRaMM025. However, PRaMM1 seems to perform better than PRaMM99.Conclusion
Differences in metabolite CVs, their CRLBs and their quantification precision were found in response to different spline node distances throughout a LCModel fit. However, only a moderate effect of the spline baseline settings was shown, and the differences were not always consistent across all metabolites and MM. Nevertheless, rather few spline nodes (PRaMM1) seem to perform slightly better than other settings. It should be noted, that the default LCModel DKNTMN setting of 0.15ppm might carry the risk of overfitting when used with a parameterized MM model.Acknowledgements
No acknowledgement found.References
1. Soher, B. J., Doraiswamy, P. M. & Charles, H. C. A review of 1H MR spectroscopy findings in Alzheimer’s disease. Neuroimaging Clin N Am 15, 847–852 (2005).
2. Emir, U. E., Tuite, P. J. & Lin O ¨ Z, G. Elevated Pontine and Putamenal GABA Levels in Mild-Moderate Parkinson Disease Detected by 7 Tesla Proton MRS. doi:10.1371/journal.pone.0030918.
3. Heckova, E. et al. Effects of different macromolecular models on reproducibility of FID-MRSI at 7T. Magn Reson Med 83, 12–21 (2020).
4. Simicic, D. et al. In vivo macromolecule signals in rat brain 1H-MR spectra at 9.4T: Parametrization, spline baseline estimation, and T2 relaxation times. Magn Reson Med 86, 2384–2401 (2021).
5. Giapitzakis, I. A., Avdievich, N. & Henning, A. Characterization of macromolecular baseline of human brain using metabolite cycled semi-LASER at 9.4T. Magn Reson Med 80, 462–473 (2018).
6. Pfeuffer, J., Tkáč, I., Provencher, S. W. & Gruetter, R. Toward an in Vivo Neurochemical Profile: Quantification of 18 Metabolites in Short-Echo-Time 1H NMR Spectra of the Rat Brain. Journal of Magnetic Resonance 141, 104–120 (1999).
7. Giapitzakis, I. A., Borbath, T., Murali-Manohar, S., Avdievich, N. & Henning, A. Investigation of the influence of macromolecules and spline baseline in the fitting model of human brain spectra at 9.4T. Magn Reson Med 81, 746–758 (2019).
8. Marjańska, M. & Terpstra, M. Influence of fitting approaches in LCModel on MRS quantification focusing on age-specific macromolecules and the spline baseline. NMR Biomed 34, 1–9 (2021).
9. Riemann, L. T. et al. Assessment of measurement precision in single-voxel spectroscopy at 7 T: Toward minimal detectable changes of metabolite concentrations in the human brain in vivo. Magn Reson Med 87, 1119–1135 (2022).
10. Rieman, Layla Tabea, Aigner, Christoph Stefan, Ellison, Stephen L.R., Brühl, Rüdiger, Mekle, Ralf, Schmitter, Sebastian, Speck, Oliver, Rose, Georg, Ittermann, Bernd, & Fillmer, A. NeuroMET - SPECIAL MRS Reproducibility [Data set]. In Magnetic Resonance in Medicine (1.0). Dataset on Zenodo (2021) doi:https://doi.org/10.5281/zenodo.5500320.
11. Mekle, R. et al. MR spectroscopy of the human brain with enhanced signal intensity at ultrashort echo times on a clinical platform at 3T and 7T. Magn Reson Med 61, 1279–1285 (2009).
12. Dell’Orco, A., Riemann, L. T., Aydin, S., Scheel, M. & Fillmer, A. Macromolecule modelling for improved metabolite quantification using very short echo time MRS at 3T: The PRaMM model. in ISMRM (2022).
13. Friedman, M. The Use of Ranks to Avoid the Assumption of Normality Implicit in the Analysis of Variance. J Am Stat Assoc 32, 675–701 (1937).
14. Wilcoxon, F. Individual Comparisons by Ranking Methods. Biometrics Bulletin 1, 80 (1945).
15. Harville, D. A. Maximum Likelihood Approaches to Variance Component Estimation and to Related Problems. J Am Stat Assoc 72, 320–338 (1977).
Figures