3430
Magnetic Resonance Spectroscopy Spectral Registration with Unsupervised Deep Learning1Department of Biomedical Engineering, Columbia University, New York, NY, United States, 2Department of Psychiatry, Columbia University, New York, NY, United States, 3Department of Neurology, Columbia University, New York, NY, United States, 4Taub Institute Research on Alzheimer's Disease and the Aging Brain, Columbia University, New York, NY, United States, 5New York State Psychiatric Institute, Columbia University, New York, NY, United States, 6Radiology and Biomedical Imaging of Biomedical Engineering, Yale University, New Haven, CT, United States, 7Mortimer B. Zuckerman Mind Brain Behavior Institute, Columbia University, New York, NY, United States
Synopsis
Keywords: Data Processing, Machine Learning/Artificial Intelligence
A deep learning-based registration method has been a successful image processing tool adopted in medical image registration but there is a lack of learning-based registration tools for spectral registration protocols. A novel CNN-based unsupervised deep learning spectral registration model was developed and trained on a simulation dataset. The model was then further evaluated on a simulated test set with more extreme conditions and on an in vivo dataset and was compared performances to published frequency-and-phase correction models. An unsupervised deep learning-based spectral registration approach was found to demonstrate state-of-the-art performance in frequency-and-phase correction.Introduction
Spectral Registration is a widely used technique to correct frequency and phase offsets. This technique is widely implemented in spectral editing software1,2,3 and applied on Magnetic Resonance Spectroscopy (MRS) data. However, due to MR being highly sensitive to scanner variabilities, frequency and phase shifts may arise, affecting data analysis. To resolve this low registration efficiency, a learning-based registration method can be considered. Deep learning has been an effective and successful image processing tool adopted in medical image registration4,5. This learning-based registration method optimizes a global functional for a dataset during training, thereby limiting time-consumption and computationally expensive optimizations during inference. In addition, a multilayer perceptron (MLP) model6 and a convolutional neural network (CNN) model7 have been recently applied to single-transient sequential frequency-and-phase correction (FPC) for edited MRS. Thus, in this study, we aim to investigate the feasibility and utility of unsupervised learning-based CNNs for spectral registration of single voxel MEGA-PRESS MRS data.Material and Methods
2.1 Simulated DatasetsMEGA-PRESS training, validation, and test transients were simulated using the FID-A toolbox as described in the previous work6,7. The training set was allocated 32,000 OFF+ON spectra, and 4,000 for both the validation and test set. The models were tested using datasets with added random Gaussian noise at SNR 20 and further challenged with SNR 2.5 + Line Broadening (LB).
2.2 In vivo Datasets
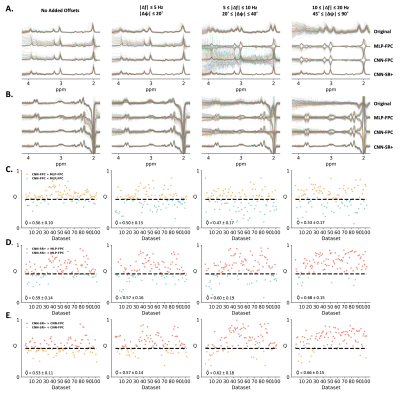
In vivo data was retrieved from the publicly available Big GABA repository8. All 101 MEGA-edited datasets from nine sites with Philips scanners were collected in total where each dataset contained 320 transients OFF+ON. The models were also evaluated on this Philips in vivo dataset with additional small offsets C1, medium offsets C2, and large offsets C3.
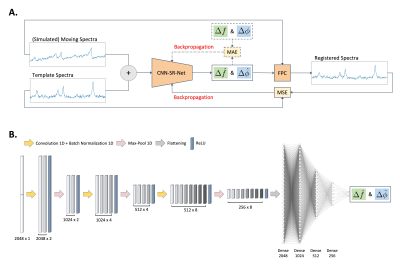
2.3 Network Architecture
A supervised and unsupervised learning loss were incorporated in our proposed model, CNN-SR, and trained on the simulation dataset to optimize the network parameters (Figure 1A). A refined model CNN-SR+ was defined by further fine-tuning the network parameters using unsupervised learning on the dataset. Our network (Figure 1B) took moving spectra and template spectra as inputs and predicted frequency and phase offsets simultaneously. An Adam optimizer9 was used to train the neural network with a 0.0001 learning rate. CNN-SR was trained for 300 epochs with a batch size of 32, and the mean absolute error was used as the loss function.
2.4 Performance Measurement
In the simulated dataset, the artificial offsets were set as the ground truth, and the mean absolute error between the ground truth and predicted value was used as the criteria to measure the network's performance. For the in vivo dataset, a Q score6 was used to determine the performance strengths of each method.
Results
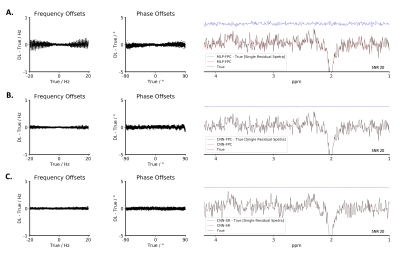
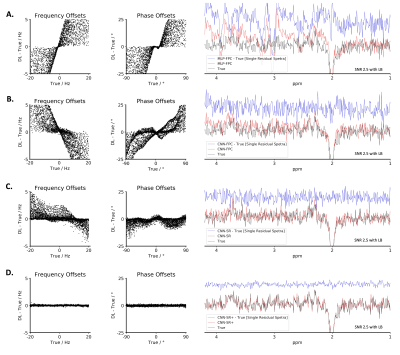
3.1. Simulated DatasetCNN-SR was capable of correcting frequency offsets (ex. Off spectra: 0.014 ± 0.010 Hz at SNR 20 and 0.678 ± 0.883 Hz at SNR 2.5 + LB) and phase offsets (ex. Off spectra: 0.104 ± 0.076° at SNR 20 and 2.367 ± 2.616° at SNR 2.5 + LB). Both these results were significantly better when compared to MLP-FPC and CNN-FPC for all conditions. Using CNN-SR+ on the SNR 2.5 + LB dataset, the performance further improved to 0.058 ± 0.050 Hz and to 0.416 ± 0.317°. Additionally, residual spectra errors in the GABA region and Glx region were found to be lower using CNN-SR in both datasets. These results can be observed from Figure 2, 3 and 4.
3.2. In vivo Dataset
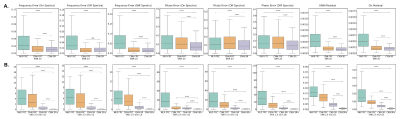
Using in vivo datasets, CNN-SR+ achieved the best performance without and with different magnitudes of additional frequency and phase offsets applied (C1, C2, and C3) followed by CNN-FPC (Figure 5).
Discussion and Conclusion
Figure 2, 3, and 4 demonstrate that when tested on simulated data (SNR 20 and SNR 2.5 + LB), MLP-FPC exhibits larger correction errors for both frequency and phase offset, and residual spectra errors (ex. GABA, Glx) followed by CNN-FPC, with both being outperformed by CNN-SR. The scatter plots and single residual spectra plots also demonstrate that CNN-SR offset predictions are more accurate than the FPC models. Moreover, in Figure 3 and 4, it can be observed that CNN-SR+ significantly improves the results. When testing on in vivo data, the utility of an unsupervised spectral registration model, CNN-SR+, demonstrates once again to have far superior performance. Figure 5 shows that Off, and Diff spectra are clearer for this model across all testing conditions, and Q scores are consistently higher.This work provides the first proof of concept of the feasibility of a CNN framework for MRS spectral registration using unsupervised learning. Despite the utility of our framework, several limitations exist such as human data being only analyzed, and only data from the MEGA-PRESS sequence being tested. Testing on animal data, and data collected from other JDE sequences (ex. MEGA-sLASER) are important variables to consider for furture exploration. Additionally, we only considered frequency and zero-order phase in this study, however, inclusion of other parameters like first-order phase, amplitude and bandwidth variance in different transients could be examined in upcoming research studies.
Acknowledgements
This study was performed at the Zuckerman Mind Brain Behavior Institute at Columbia University and Columbia MR Research Center site.References
1. Liu C, Ma D. J., et al (2021) JET - A Matlab toolkit for automated J-difference-edited MR spectra processing of in vivo mouse MEGA-PRESS study at 9.4T. In Joint Annual Meeting ISMRM & SMRT 2021 (Vancouver, Canada).
2. Near J, Edden R, Evans CJ, Paquin R, Harris A, Jezzard P. Frequency and phase drift correction of magnetic resonance spectroscopy data by spectral registration in the time domain. Magn Reson Med. 2015;73:44-50.
3. Oeltzschner G, Zöllner HJ, Hui SCN, et al. Open-source processing, reconstruction and estimation of magnetic resonance spectroscopy data. J Neurosci Methods. 2020;343:108827.
4. Fu Y, Lei Y, Wang T, Higgins K, Bradley JD, Curran WJ, Liu T, Yang X. LungRegNet: An unsupervised deformable image registration method for 4D-CT lung. Med Phys. 2020 Apr;47(4):1763-1774. doi: 10.1002/mp.14065. Epub 2020 Feb 26. PMID: 32017141; PMCID: PMC7165051.
5. Chen, J., Frey, E.C., Du, Y. (2022). Unsupervised Learning of Diffeomorphic Image Registration via TransMorph. In: Hering, A., Schnabel, J., Zhang, M., Ferrante, E., Heinrich, M., Rueckert, D. (eds) Biomedical Image Registration. WBIR 2022. Lecture Notes in Computer Science, vol 13386. Springer, Cham.
6. Tapper S, Mikkelsen M, Dewey BE, Zöllner HJ, Hui SCN, Oeltzschner G, Edden RAE. Frequency and phase correction of J-difference edited MR spectra using deep learning. Magn Reson Med. 2021 Apr;85(4):1755-1765. doi: 10.1002/mrm.28525. Epub 2020 Nov 18. PMID: 33210342.
7. Ma DJ, Le HA, Ye Y, Laine AF, Lieberman JA, Rothman DL, Small SA, Guo J. MR spectroscopy frequency and phase correction using convolutional neural networks. Magn Reson Med. 2022 Apr;87(4):1700-1710. doi: 10.1002/mrm.29103. Epub 2021 Dec 21. PMID: 34931715.
8. Mikkelsen M, Barker PB, Bhattacharyya PK, et al. Big GABA: Edited MR spectroscopy at 24 research sites. Neuroimage. 2017;159:32-45.
9. D. Kingma and J. Ba. Adam: A method for stochastic optimization”. arXiv 2014, arXiv:1412.6980.
Figures