3427
Streak Artifact Reduction using Convolutional Neural Network: Clinical Feasibility Study in Liver MR Imaging1Hamamatsu University School of Medicine, Hamamatsu, Japan
Synopsis
Keywords: Artifacts, Machine Learning/Artificial Intelligence, Liver, Motion Correction
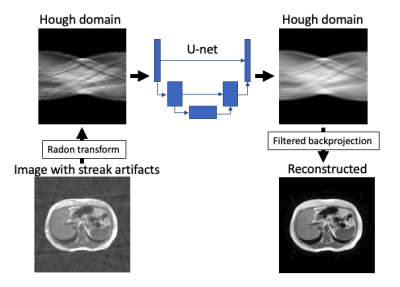
Radial sampling enables free free-breathing abdominal MR imaging. Meanwhile, while it suffers from streak artifacts. We propose streak artifact reduction using convolutional neural network (SARC) which utilize Hough domain. In Hough domain, a streak becomes like a dot which is more localized compared with image domain. The network was trained in end-to-end manner. The SARC was show better image quality in objective image quality metrics and visual evaluation by a radiologist. SARC showed feasibility for clinical MR imaging.Introduction
Liver MRI examination plays an important role in diagnosis of liver tumors. It is utilized in LI-RADS (Liver Reporting and Data System) to diagnose hepatocellular carcinoma which is the most common etiology of liver cancer.1 However, liver MRI is one of the most difficult examination due to respiratory motion of the abdominal organs and severe motion artifacts.Free breathing acquisition using radial sampling is a promising technique to reduce motion artifact2, while it suffers some streak artifacts. To handle this issue, we propose streak artifact reduction using convolutional neural network (SARC). We demonstrate performance of SARC compared to conventional technique and clinical feasibility.
Methods
For network training and evaluation, we used 40 cases of 2D-T1 weighted dual echo imaging from CHAOS public dataset.3 A total of 1570 images were used for network training and 1100 images were used for evaluation. For training and evaluation, all images were resized 256 x 256 matrix and undersampled with 80 spokes (10% of Nyquist sampling).The network architecture of proposed convolutional neural network (SARC) consists of 3 steps (Figure 1). First, an input image is converted to Hough domain using Radon transform. Second, a U-net based network is applied to the Hough domain to remove artifact which reflects streak artifact in image domain.4 Finally, filtered backprojection to image domain is performed. Training parameters were used as follows: end-to-end training; loss function, mean squared error; optimizer, SGD; learning rate, 1.0e-4. For comparison, the SARC and previously reported deep learning-based respiratory motion artifact reduction (MARC) were applied to undersampled images.5 Undersampled images were also evaluated as baseline. The image quality was evaluated objectively using structure similarity index (SSIM). To estimate clinical feasibility, visual evaluation by a radiologist was also performed using 5-point scale (1, not diagnostic; 3, fair; 5, excellent) in terms of streak artifact, conspicuity of intrahepatic vessels, and overall image quality. Wilcoxon signed rank test was used to compare these metrics. Bonferroni correction was applied to compensate multiple comparison. A p-value of 0.05 was considered as significant. All statistical analyses were performed using R (2022, version 4.2.0, R: A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria).
Results
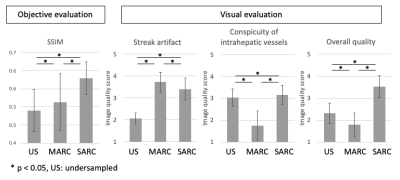
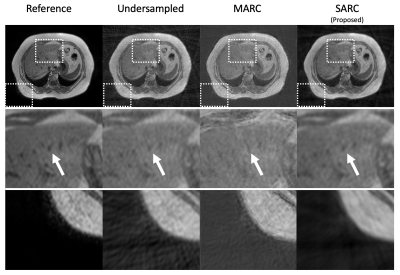
SSIM of the three methods were 0.490±0.059, 0.513±0.078, and 0.579±0.046 (mean±standard deviation; undersampled, MARC, and SARC, respectively). For streak artifact reduction, MARC showed the best performance (P < 0.05) but the difference between MARC and SARC was small. Meanwhile MARC showed the worse for conspicuity of intrahepatic vessels (P < 0.05). For overall image quality, SARC showed the best score (P < 0.05). The details were showed in Figure 2. The representative case was showed in Figure 3.Discussion
MARC showed the best performance for reducing streak artifacts, while degrade conspicuity of intrahepatic vessels. We speculate it is not easy to discriminate vessel structure from streak artifact because both of them have linear shape. SARC showed the best overall image quality preserving conspicuity of intrahepatic vessels. This suggests higher specificity of streak artifact in Hough domain which is utilized in SARC. These results showed clinical feasibility of SARC to reduce streak artifact. The major limitation of this study is that only basic clinical image quality was evaluated, and hepatic lesions were not evaluated.Conclusion
SARC showed better performance of streak artifact reduction compared with conventional technique preserving conspicuity of intrahepatic vessels.Acknowledgements
No acknowledgement found.References
1. Chernyak V, Fowler KJ, Kamaya A, et al.: Liver Imaging Reporting and Data System (LI-RADS) Version 2018: Imaging of Hepatocellular Carcinoma in At-Risk Patients. Radiology 2018; 289:816–830.
2. Ichikawa S, Motosugi U, Kromrey M-L, et al.: Utility of Stack-of-stars Acquisition for Hepatobiliary Phase Imaging without Breath-holding. Magn Reson Med Sci 2020; 19:99–107.
3. Kavur AE, Selver MA, Dicle O, Barış M, Gezer NS: CHAOS - Combined (CT-MR) Healthy Abdominal Organ Segmentation Challenge Data. 2019.
4. Ronneberger O, Fischer P, Brox T: U-Net: Convolutional Networks for Biomedical Image Segmentation. In Med Image Comput Comput-Assist Interv – MICCAI 2015. Volume 9351. Edited by Navab N, Hornegger J, Wells WM, Frangi AF. Cham: Springer International Publishing; 2015:234–241.
5. Tamada D, Kromrey ML, Ichikawa S, Onishi H, Motosugi U: Motion artifact reduction using a convolutional neural network for dynamic contrast enhanced mr imaging of the liver. Magn Reson Med Sci 2020; 19:64–76.
Figures