3308
Modality-adaptive Network Learning for Brain Tumor Segmentation with Incomplete Multi-modal MRI Data1Paul C. Lauterbur Research Center for Biomedical Imaging, Shenzhen Institutes of Advanced Technology, Chinese Academy of Sciences, Shenzhen, China, 2School of Computer and Artificial Intelligence, Zhengzhou University, Zhengzhou, China, 3Peng Cheng Laboratory, Shenzhen, China, 4Guangdong Provincial Key Laboratory of Artificial Intelligence in Medical Image Analysis and Application, Guangdong Provincial People’s Hospital, Guangdong Academy of Medical Sciences, Guangzhou, China
Synopsis
Keywords: Segmentation, Brain
Automated brain tumor segmentation with multi-modal magnetic resonance imaging (MRI) data is crucial for brain cancer diagnosis. Nevertheless, in clinical applications, it is difficult to guarantee that complete multi-modal MRI data are available due to different imaging protocols and inevitable data corruption. A large test time performance drop could happen. Here, we design a modality-adaptive network learning method to extract common representations from different modalities and make our trained model applicable to different data-missing scenarios. Experiments on an open-source dataset demonstrate that our method can reduce the dependence of deep learning-based segmentation methods on the integrity of input data.Introduction
Segmenting brain tumor regions in multi-contrast MRI scans is important for pre-operative treatment planning and postoperative monitoring of patients with brain tumors. Manually delineating the tumor boundaries is time-consuming and error-prone owning to its structural complexities and histologic differences1. Many automated multi-modal MR image segmentation methods have been developed, especially those based on deep learning (DL)2-5. However, most of the existing DL-based methods rely heavily on the integrity of the multi-modal MRI data. In deployment, their performances can deviate largely from the expectation when facing incomplete multi-modal MR data. Although intuitively we can train individual segmentation models for any possible modality missing condition to avoid the domain shift between training and inference data, it’s time-consuming and less efficient. Some methods leveraged support vector machines and random forest together with a regression strategy to help synthesize the missing data66,7. Recently, Havaei M et al.8 and Dorent R et al.9 utilized the power of Variational Auto Encoders (VAE) to search the modality-shared latent space. Despite the achieved promising results, further attempts are still needed to effectively extract the common features from the multi-modal MR data. Here, we propose a modality-adaptive network which aims to build an explicit relationship between the common multi-modal features and the modality-shared features through transfer learning. To further constrain the knowledge transfer process, a loss is designed for different decoding stages to preserve the integrity of multi-modal information and enhance the overall segmentation performance.Method
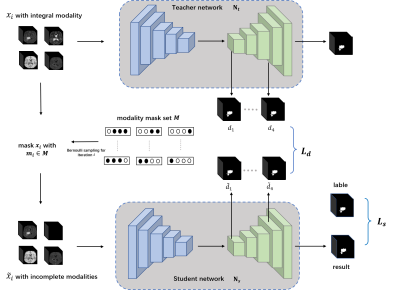
Our proposed modality-adaptive network is shown in Fig.1. It includes a teacher network $$$N_t$$$ and a student network $$$N_s$$$. Both networks employ a U-Net10 backbone with small modifications. The learning process of our proposed modality-adaptive network can be divided into two parts: 1) Pretrain $$$N_t$$$ using a complete multi-modal MRI data set $$$X$$$. 2) For $$$N_s$$$ we use modality-incomplete counterpart $$$\widetilde{X}$$$ of $$$X$$$ to implement modality-shared knowledge distillation training guided by $$$N_t$$$. Specifically, during every iteration $$$i$$$, the training data batch $$$\widetilde{x}_i$$$ of $$$N_s$$$ is generated from multiplying a modality mask, which follows a Bernoulli distribution $$$B(1,0.5)$$$, with the modality encoding. Incomplete training data generation in this manner empowers $$$N_s$$$ to traverse samples of different modality-missing conditions and adaptively capture the modality-invariant information for the segmentation task. To help transfer as much as possible the modality-shared information from $$$N_t$$$ to $$$N_s$$$, we further introduce a loss $$$L_d$$$ leveraging deep supervision11. Let $$$d_i$$$ and $$$\widetilde{d}_i$$$ denote the output segmentation maps generated from the decoder level $$$i$$$ by the deep supervision blocks for $$$N_t$$$ and $$$N_s$$$ respectively, we use the absolute difference to constrain them. Given a batch size of $$$N$$$, $$$L_d$$$ is defined as: $$L_d=\frac{1}{N}\sum_i\left|d_i - \widetilde{d}_i\right|$$To enhance the final segmentation performance of Ns, we use Dice loss for the output of the final decoder stage:$$L_s=1-\frac{1}{N}\sum_{j}\frac{M_j*P_j+\varepsilon}{M_j^2+P_j^2+\varepsilon}$$where $$$M_j$$$ refers to the output probability after sigmoid activation. $$$P_j$$$ refers to the ground truth label at output channel $$$j$$$. $$$\varepsilon$$$ is a smoothing factor. The overall objective function for the proposed modality-adaptive network can be described as:$$\mathop{arg\min}\left(L_d+L_s\right)$$
Results and Discussion
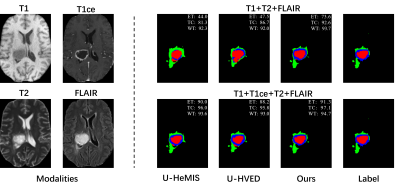
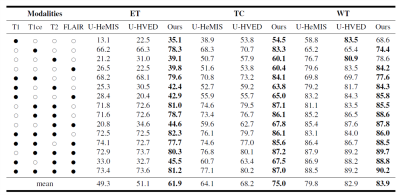
Effectiveness of the proposed network has been evaluated on an open-source brain tumor segmentation dataset, BraTs202012-14. BraTs2020 includes four MRI modalities: T1, T1ce, T2, and FLAIR. The tumor area in each case is divided into the GD-enhancing tumor (ET), the peritumoral edema (ED), and the necrotic and non-enhancing tumor core (NCR/NET) with radiologists approved annotations. These three brain tumor categories can be further combined into the enhancing tumor (ET), the tumor core (TC=ET+NCR), and the whole tumor (WT=ET+NCR+ED). The segmentation task of the challenge is to delineate ET, TC, and WT.Quantitative results of different methods are listed in Table 1. Two comparison methods are adopted, U-HeMIS and U-HVED7,8. It’s worth noting that our method achieves the best performance in all 15 scenarios, which include various incomplete multi-modal MRI data conditions as well as the complete data condition. Fig. 2 plots example segmentation maps of different methods in two scenarios. It can be observed in Fig. 2 that our method is able to clearly delineate the tumor core boundary even when T1 scans are missing.Conclusion
In this work, we propose a modality-adaptive deep learning network for brain tumor segmentation, which addresses the incomplete multi-modal MRI data problem faced during model deployment. With our proposed distillation training method, the proposed network can extract rich modality-shared features and adapt to different modality-missing scenarios. Experimental results on BraTs2020 verify that our method has a high potential in mitigating the performance degradation issue encountered by existing deep learning-based segmentation models when incomplete multi-modal MR data are provided for inference, which can be very helpful for clinical applications by automating MR image segmentation.Acknowledgements
This research was partly supported by Scientific and Technical Innovation 2030-"New Generation Artificial Intelligence" Project (2020AAA0104100, 2020AAA0104105), the National Natural Science Foundation of China (61871371,62222118,U22A2040), Guangdong Provincial Key Laboratory of Artificial Intelligence in Medical Image Analysis and Application(No. 2022B1212010011), the Basic Research Program of Shenzhen (JCYJ20180507182400762), Shenzhen Science and Technology Program (Grant No. RCYX20210706092104034), AND Youth Innovation Promotion Association Program of Chinese Academy of Sciences (2019351).References
1. Bakas S, Akbari H, Sotiras A, et al. Advancing the cancer genome atlas glioma MRI collections with expert segmentation labels and radiomic features. Scientific data, 2017, 4(1): 1-13.
2. Zhou Y, Huang W, Dong P, et al. D-UNet: a dimension-fusion U shape network for chronic stroke lesion segmentation. IEEE/ACM transactions on computational biology and bioinformatics, 2019, 18(3): 940-950.
3. Wang S, Li C, Wang R, et al. Annotation-efficient deep learning for automatic medical image segmentation. Nature communications, 2021, 12(1): 1-13.
4. Pusterla O, Heule R, Santini F, et al. MRI lung lobe segmentation of pediatric cystic fibrosis patients using a neural network trained with publicly accessible CT datasets. Joint Annual Meeting ISMRM-ESMRMB & ISMRT 31st Annual Meeting (ISMRM 2022). 2022.
5. Wech T, Ankenbrand M J, Bley T A, et al. A data‐driven semantic segmentation model for direct cardiac functional analysis based on undersampled radial MR cine series[J]. Magnetic Resonance in Medicine, 2022, 87(2): 972-983.
6. Jog A, Carass A, Roy S, et al. Random forest regression for magnetic resonance image synthesis. Medical image analysis, 2017, 35: 475-488.
7. Hofmann M, Steinke F, Scheel V, et al. MRI-based attenuation correction for PET/MRI: a novel approach combining pattern recognition and atlas registration. Journal of nuclear medicine, 2008, 49(11): 1875-1883.
8. Havaei M, Guizard N, Chapados N, et al. Hemis: Hetero-modal image segmentation. International Conference on Medical Image Computing and Computer-Assisted Intervention. Springer, Cham, 2016: 469-477.
9. Dorent R, Joutard S, Modat M, et al. Hetero-modal variational encoder-decoder for joint modality completion and segmentation. International Conference on Medical Image Computing and Computer-Assisted Intervention. Springer, Cham, 2019: 74-82.
10. Çiçek Ö, Abdulkadir A, Lienkamp S S, et al. 3D U-Net: learning dense volumetric segmentation from sparse annotation[C]. International conference on medical image computing and computer-assisted intervention. Springer, Cham, 2016: 424-432.
11. Wang L, Lee C Y, Tu Z, et al. Training deeper convolutional networks with deep supervision. arXiv preprint arXiv:1505.02496, 2015.
12. Menze B H, Jakab A, Bauer S, et al. The multimodal brain tumor image segmentation benchmark (BRATS). IEEE transactions on medical imaging, 2014, 34(10): 1993-2024.
13. Bakas S, Akbari H, Sotiras A, et al. Advancing the cancer genome atlas glioma MRI collections with expert segmentation labels and radiomic features. Scientific data, 2017, 4(1): 1-13.
14. Bakas S, Reyes M, Jakab A, et al. Identifying the best machine learning algorithms for brain tumor segmentation, progression assessment, and overall survival prediction in the BRATS challenge. arXiv preprint arXiv:1811.02629, 2018.
Figures