3301
Fast 3D-P31-MRSI Using Custom Rosette Petal Trajectory at 3T with 4x Accelerated Compressed Sensing
Nicholas Farley1, Brian Bozymski1, Ulrike Dydak1, and Uzay Emir1
1Radiological Health Science, Purdue University, West Lafayette, IN, United States
1Radiological Health Science, Purdue University, West Lafayette, IN, United States
Synopsis
Keywords: Gray Matter, Brain
To develop a reasonably fast P-31 3D MRSI sequence utilizing Rosette-Petal trajectories in K-space and compressed sensing reconstruction to acquire 1 mL voxel resolution maps in under 10 minutes with a magnetic field strength of only 3 TeslaSynopsis
To develop a reasonably fast P-31 3D MRSI sequence utilizing Rosette-Petal trajectories in K-space and compressed sensing reconstruction to acquire 1 mL voxel resolution maps in under 10 minutes with a magnetic field strength of only 3 Tesla.Introduction
Phosphorus Magnetic Resonance Spectroscopy (31-P MRS) is a useful method for non-invasive-in-vivo quantification of phosphorus containing metabolites within the human brain. Amongst these metabolites are Adenosine Triphosphate (ATP) and Phosphocreatine (PCr) which constitute two pools of high-energy phosphates (HEP) and thereby serve as useful indicators for local energy metabolism. However, 31-P MRS suffers from poor signal response relative to 1-H MRS, which thereby necessitates longer scan durations or lower-spatial resolutions to compensate. This problem is only magnified in Multi-Voxel Spectroscopy (31-P MRSI) where the desire for multiple high-resolution voxels leads to unacceptably long scan durations. Since Phosphate containing metabolites have been observed to vary significantly between different brain regions (Dudley et al.), a reasonably fast 3D-31P-MRSI pulse sequence would be extremely useful for investigating the heterogeneous energy metabolism of the human brain. Over the previous two decades multiple advances have been made in sequence development. One of the earliest attempts in 2001 managed 6.3 mL resolution in 46 minutes at 4T (Hetherington et al.) then in 2021 another group reported 0.59 mL resolution with a total scan time of 74 minutes using a 9.4T scanner (Ruhm et al.). Around the same time another group used modified Concentric Ring Trajectories to spherically sample k-space and obtain sufficiently high SNR in 2.5 minutes with a resolution of 6.7 mL (Clarke et al.). For this abstract, our group proposes a 31-P MRSI sequence utilizing Rosette-Petal shaped 3D k-space acquisition to achieve 1 mL isotropic resolution (10x10x10mm^3) over a FOV of 480x480x480mm^3 with a total scanning duration of 10 minutes using a Siemens Prisma 3T scanner through the implementation of compressed sensing reconstruction for x4 acceleration.Methods
Data was acquired from the 3T Siemens Prisma Scanner in Purdue University’s MRI facility. Only one set of data was acquired for this study. Subject was positioned supine head-first and measured with a previously custom-built, dual-tuned 1H/31P Tx/Rx 8-channel volume coil. A custom rosette trajectory (Shen et al.) was used with an isotropic FOV = (480mm)^3 and an imaging matrix grid of 48x48x48 voxels. The spectral bandwidth was 2.2 kHz, with 256 points in the temporal dimension, a TE = 70 us, a TR = 350 ms, and a total of 1444 Petals. Total scan duration = 30 minutes after taking three averages. With Compressed Sensing Reconstruction we could accelerate data acquisition by a factor of 4 to bring our accelerated total scan time down to ~8 minutes. Anatomical data was acquired through an Ultrashort Echo Time Magnetization Transfer (UTE-MT) sequence which uses a pair of adiabatic hyperbolic secant (sech) 180 degree pulses with a pulse duration of 24ms, a pulse bandwidth of 1kHz, an offset frequency of ~1300 Hz from water. Each sech pulse was repeated an additional 9 times for averaging. Total scan duration = 12 minutes. Voxel-wise output spectra were post-processed in Matlab to obtain concentration and Cramer-Rao Lower Bound (Standard Deviation) values for ATP, PCr, and inorganic phosphate (Pi). FSLeyes was used to co-register metabolite maps with the anatomical image.Results and Discussion
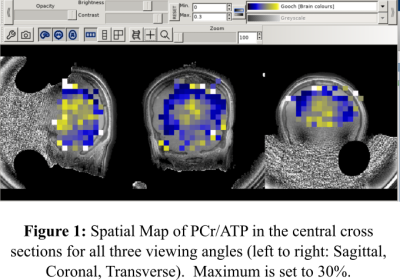
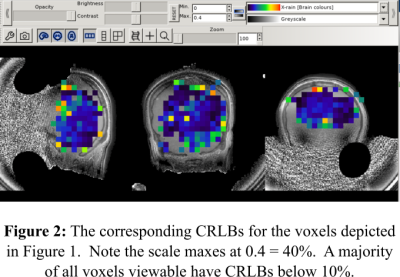
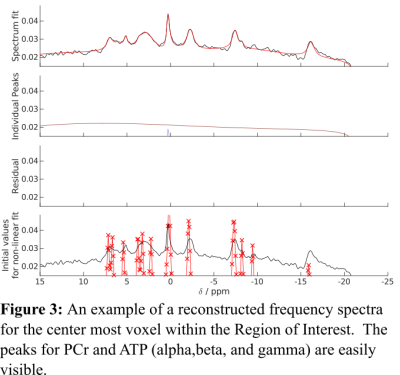
The center slice of the concentration and CRLB maps for PCr/TP is displayed in Figures 1 and 2. Additionally, Figure 3 shows an example of acquired P-31 spectra in the center of the FOV. Our results demonstrate the feasibility of acquiring 31-P MRSI data in under 10 minutes with only magnetic field strength of 3 Tesla; but more importantly, they hint to the possibility of faster 31-P MRSI data acquisition through the manipulation of the k-space trajectory. In the future our group plans to continue investigating alternative k-space trajectories.Acknowledgements
No acknowledgement found.References
Clarke, William T., et al. "Three‐dimensional, 2.5‐minute, 7T phosphorus magnetic resonance spectroscopic imaging of the human heart using concentric rings." NMR in Biomedicine (2022): e4813. Dudley, J., et al. "Tissue dependent metabolism in the human brain suggested by quantitativephosphorus-31 MRSI." J Spectrosc Dyn 4 (2014): 19. Hetherington, H. P., et al. "Quantitative 31P spectroscopic imaging of human brain at 4 Tesla: assessment of gray and white matter differences of phosphocreatine and ATP." Magnetic Resonance in Medicine: An Official Journal of the International Society for Magnetic Resonance in Medicine 45.1 (2001): 46-52. Ruhm, Loreen, et al. "3D 31P MRSI of the human brain at 9.4 Tesla: Optimization and quantitative analysis of metabolic images." Magnetic resonance in medicine 86.5 (2021): 2368-2383. Shen, Xin, et al. "Ultra-short T2 components imaging of the whole brain using 3D dual-echo UTE MRI with rosette k-space pattern." Magnetic Resonance in Medicine (2022).
DOI: https://doi.org/10.58530/2023/3301