3281
Sequence optimization for saturation transfer MR fingerprinting1Russell H. Morgan Department of Radiology and Radiological Science, Johns Hopkins University, Baltimore, MD, United States
Synopsis
Keywords: CEST & MT, CEST & MT
An optimal saturation transfer MR fingerprinting (ST-MRF) acquisition schedule is critical for efficient and accurate tissue parameter mapping. To optimize RF saturation-encoded MRF acquisitions to a minimal number of saturation scan parameters for magnetization transfer contrast (MTC) and chemical exchange saturation transfer (CEST) parameter determination, we developed an optimization framework using a support vector regression-recursive feature elimination (SVR-RFE) method. Bloch simulations and in vivo studies showed that the proposed optimization method outperformed the quantification accuracy compared to existing methods. The SVR-RFE-based optimization method allowed us to reduce scan time by ~50% without sacrificing reconstruction accuracy.Introduction
Conventional magnetization transfer contrast (MTC) and chemical exchange saturation transfer (CEST) MRI approaches acquire a series of images at multiple frequency offsets1-3. Sometimes, a highly sampled Z-spectra acquisition is required to correct field inhomogeneity and isolate multiple CEST effects, which is time-consuming. A recent study showed that a linear projection method with L1-regularization can reduce frequency offset acquisition schedules, thus reducing total acquisition time4. Another optimization approach using a deep-learning neural network was introduced for quantitative MTC MR fingerprinting (MRF), which improved reconstruction accuracy and acquisition efficiency5,6. However, a multi-pool exchange model increases the number of tissue parameters to be determined and computational complexity. Here, we developed a support vector regression-recursive feature elimination (SVR-RFE) method to optimize saturation transfer MRF (ST-MRF) with a three-pool exchange model (free water, semisolid macromolecule, and amide protons), which was compared with least absolute shrinkage and selection operator (LASSO)7,8. LASSO typically uses least-squared regression with L1 regularization and minimizes the distance from each observation, whereas SVR maximizes the possible margin between the targets. We accelerated imaging acquisition by reducing redundant information from dynamic ST-MRF images.Methods
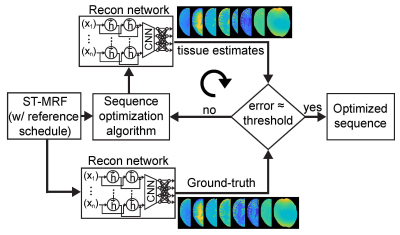
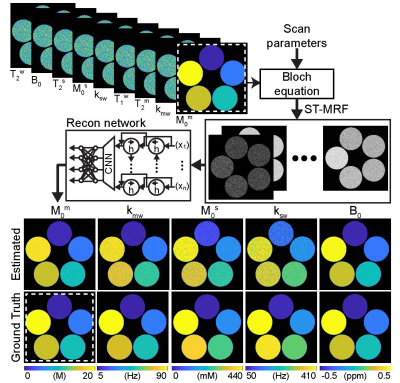
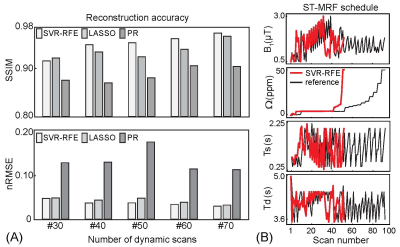
Training dataset was synthesized by solving three-pool Bloch equations with a pre-defined ST-MRF schedule with sufficient dynamic scans (93 dynamic scans, a reference schedule) and tissue parameter combinations: free water T1 (T1w) and T2 (T2w) relaxation times, semisolid macromolecular proton exchange rate (kmw), concentration (M0m), and T2 relaxation times (T2m), amide proton exchange rate (ksw), concentration (M0s), and T2 relaxation times (T2s), and B0 shift parameters. The importance of each dynamic scan parameter was measured by recursively removal of samples from the reference ST-MRF signal. The ST-MRF schedule with various numbers of dynamic scans (#30, #40, #50, #60, and #70) were obtained from the ranked reference dynamic schedule. For tissue quantification, a recurrent neural network was trained with the reference schedule, pseudorandomized (PR) schedule, LASSO-optimized schedule, and SVR-RFE-optimized schedule (Fig. 1). In the digital phantom study, as illustrated in Fig. 2, the quantification accuracy was evaluated by calculating normalized root mean square error (nRMSE) values between ground-truths and estimated tissue parameters. Nine normal subjects were recruited for in vivo study. All subjects were examined with the approval of the institutional ethics committee, and written informed consent was obtained prior to the study. 3D ST-MRF images were acquired from a multi-shot TSE sequence with fourfold (2 x 2) compressed sensing acceleration in the Ky-Kz plane at 3T MRI. Dynamic ST-MRF images were acquired with varied frequency offsets (Ω), RF saturation strengths (B1), RF saturation times (Ts), and relaxation delay times (Td).Results and Discussion
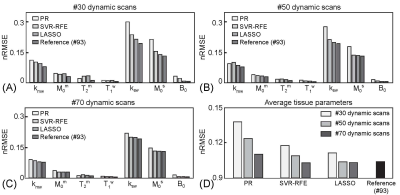
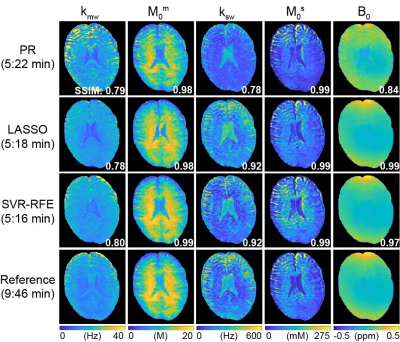
First, the SVR-RFE-based optimization technique was evaluated on digital phantoms (Figs. 2 and 3). As expected, the reference schedule with 93 dynamic scans had the lowest nRMSE, while the pseudorandomized schedule had the highest nRMSE. The optimization methods were compared against each tissue parameter and quantification accuracy was reported in Fig. 3. Overall, a reduction of the number of dynamic scans resulted in increased nRMSE. Nevertheless, both LASSO and SVM-RFE still showed a high degree of accuracy in the quantification of tissue parameters. The in vivo tissue parameter maps reconstructed from various ST-MRF schedules are shown in Fig. 4. Overall, the structural similarity index measure (SSIM) values of the SVR-RFE were higher than those of the LASSO optimization method, while the nRMSE values of the SVR-RFE were lower than those of the LASSO optimization method in in vivo experiments (n=9) (Fig. 5). We observed higher performance of SVR-RFE on in vivo data presumably because SVR calculating the support vectors lies on the edge of each class (tissue parameter) to make decision and hence, reduces the risk of overfitting on unseen data. With the SVR-RFE optimization algorithm, 50 dynamic scans provided the best trade-off between quantification accuracy (nRMSE <0.05 and SSIM > 0.95) and scan efficiency (reduced total scan time to ~50%).Conclusions
We proposed an optimization framework to improve quantification accuracy and efficiency for ST-MRF. The SVR-RFE-based sequence optimization was tested with digital phantoms and demonstrated on healthy volunteer human brains. The optimized ST-MRF framework could provide quantitative MTC and CEST parameter mappings within a clinically feasible scan time.Acknowledgements
This work was supported in part by grants from the National Institutes of Health.References
1. Balaban RS, Ceckler TL. Magnetization transfer contrast in magnetic resonance imaging. Magn Reson Q 1992;8:116-137.
2. Ward KM, Aletras AH, Balaban RS. A new class of contrast agents for MRI based on proton chemical exchange dependent saturation transfer (CEST). J Magn Reson 2000;143:79-87.
3. Wolff SD, Balaban RS. Magnetization transfer contrast (MTC) and tissue water proton relaxation in vivo. Magn Reson Med 1989;10:135-144.
4. Glang F, Fabian MS, German A, Khakzar KM, Mennecke A, Liebert A, Herz K, Liebig P, Kasper BS, Schmidt M, Zuazua E, Nagel AM, Laun FB, Dorfler A, Scheffler K, Zaiss M. Linear projection-based chemical exchange saturation transfer parameter estimation. NMR Biomed 2022:e4697.
5. Kang B, Kim B, Park H, Heo HY. Learning-based optimization of acquisition schedule for magnetization transfer contrast MR fingerprinting. NMR Biomed 2022;35(5):e4662.
6. Perlman O, Farrar CT, Heo HY. MR fingerprinting for semisolid magnetization transfer and chemical exchange saturation transfer quantification. NMR Biomed 2022:e4710.
7. Tibshirani R. Regression shrinkage and selection via the Lasso. J Roy Stat Soc B Met 1996;58(1):267-288.
8. Choi H, Yeo D, Kwon S, Kim Y. Gene selection and prediction for cancer classification using support vector machines with a reject option. Comput Stat Data An 2011;55(5):1897-1908.
Figures