3247
Automated brain QSM computation pipeline deployed in the European Open Science Cloud1Neurospin, Paris, France, 2Ventio, Marseille, France
Synopsis
Keywords: Data Processing, Quantitative Susceptibility mapping, Cloud-computing
Introduction of a pipeline for computing the Quantitative Susceptibility Mapping (QSM) from non-optimal MRI data in a secure cloud-infrastructure. The pipeline allows the computation of clean and exploitable QSM, thanks to a phase pre-processing to reduce the presence of artifacts in the 7T MRI data.Introduction
Iron accumulates in the brain over time, and an abnormal load is related to neurodegenerescence [1]. A quantitative measure of the iron load could then be used as a biomarker for evaluating the healthy brain aging as well as the pathologies severity [1][2]. QSM provides an iron load quantitative metric [3], and by means of it the accumulation of iron has been analyzed, mostly at 3T [1]. Ultra-high magnetic fields provide higher sensibility and resolution [4], but they are more vulnerable to effects from strong field variations due to air/tissue interface [2]. Also, an improper coil combination can generate artifacts in the phase reconstruction, leading to unexploitable QSM (Fig 1A). There are some available software that allow to reconstruct QSM from 3D multi-echo gradient echo (MGRE). They are mostly adapted to process data at 3T and only few offer an automatic user-independent pipeline [5]. Moreover they do not provide a means to overcome the possible artifacts in the input data. Here we propose a pipeline for obtaining QSM from 3D MGRE DICOM data without the need of in-house powerful calculation machines, since the computation takes place in a ISO27001-certified Openstack cloud infrastructure (de.NBI Clouda). It considers a phase pre-processing to reduce the presence of artifacts (from the background field and/or coil combination), and use the filtered field for the MEDI algorithm [6]. The pipeline was conceived as part of the QSM4SENIORb study, that aims at studying the accumulation of iron in a longitudinal way by using QSM in the SENIOR database [7]. We take advantage of these high resolution data for testing the pipeline.Methods
(I) Participants: 84 volunteers were selected from the SENIOR database [7].(II) Image acquisition: The MRI data was acquired at NeuroSpincd (France) on a Magnetom 7 Tesla scanner (Siemens Healthineers, Germany) 1Tx/32Rx Nova Medical head coil. MGRE was performed (TA=9:48 min, FoV=256 mm, voxel size=0.8 mm isotropic, TR=37 ms, TE=1.68 ms, TE=3.05 ms, number of echoes=10, flip angle=30°, acceleration factor GRAPPA=3, 196 sagittal partitions, bandwidth=740 Hz/px, monopolar readouts) and reconstructed using Virtual Coil Combination (VCC) [8]. The T1-weighted MP2RAGE was also acquired (TR= 6000 ms; TE=2.96 ms; voxel size=0.75 mm isotropic).
(III) Cloud computing: A virtual machine (14 cores, 32 Go RAM and 200 Go volume) was deployed and configured automatically in the de.NBI cloud. Imaging data were encrypted on transit and at rest.
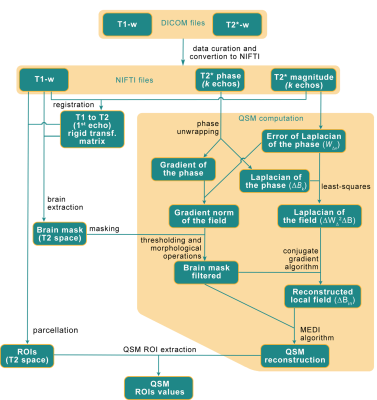
(IV) Data processing: The method consists in pre-filtering the phase data from the MGRE acquisition by using the magnitude and phase from the ten available echoes. It is based on the method described in [9] and illustrated in Fig 2. To isolate the brain internal field variations (ΔBin) the conjugate gradients algorithm of the normal equation ΔWΔ2ΔBin = ΔWΔ2ΔB is calculated, where WΔ is a diagonal matrix weighting each estimation of the Laplacian by the inverse of its error standard deviation. The unwrapping is done by forcing the point by point difference between ]-π, π] when calculating ΔB using the modulo function. The Laplacian of the field is calculated from the Laplacian of the phase for each echo k (ΔBk) and its error (WΔK) combined in the least-squares sense. The gradient norm of the field is also computed similarly.
A mask for excluding voxels suffering from spatial deformation (leading to a Bin that cannot be correctly measured) is also computed from the gradient norm of the field by applying a whole-brain mask (computed using ANTs from the T1-weighted and registered by a rigid transformation to the T2 space) over it, followed by a thresholding and morphological operations (opening, connected components and closing). Finally, the conjugate gradient algorithm is used to compute from the normal equation (stopped at maximum iterations: 512 or the relative norm less than 10-3).
Results and Discussions
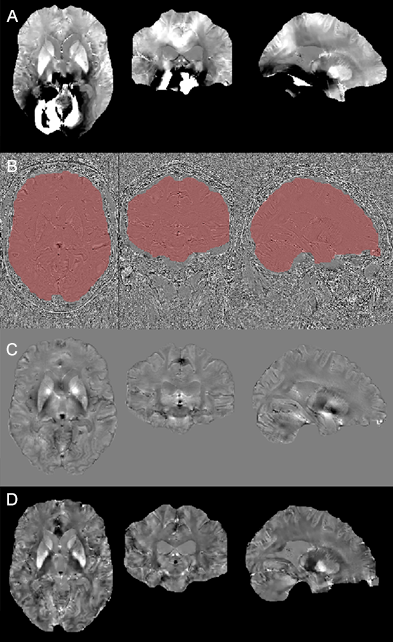
Figure 1 shows an example of a cloud-computed QSM from the reconstructed unwrapped filtered phase and the customized mask (D). The refinement of the brain mask (B) allows to remove from the QSM computation some unreliable voxel values. The computation of a filtered phase map (C) helps overcoming the presence of artifacts such as open-ended fringe lines arising from a poor coil combination, providing a cleaner input into the MEDI algorithm, obtaining QSM with reduced artifacts (D). The pre-processing pipeline may be used in the presence of phase errors stemming from reconstruction issues. The implementation of the pipeline in a virtual infrastructure demonstrates the capacity of remote processing of research data in secure environments. This offers the possibility of schedule the analyze of the whole cohort importing into the servers the DICOM data, run the pipeline and then export the results.Conclusions
We present the preliminary results for a cloud-based automatic brain QSM computation, showing the ability of our pipeline to recover QSM information from non-optimal data, thanks to the field pre-processing steps for reducing the presence of artifacts in the input phase data. Although the results are preliminary, the method is promising because it allowed us to retrieve the information from the available data, without the need for powerful in-house calculation machines. Its automatic application to the SENIOR cohort will enable the incorporation of iron load quantification as a valuable biomarker to the database.Acknowledgements
a This work was supported by the BMBF-funded de.NBI Cloud within the German Network for Bioinformatics Infrastructure (de.NBI) (031A532B, 031A533A, 031A533B, 031A534A, 031A535A, 031A537A, 031A537B, 031A537C, 031A537D, 031A538A).
b This project has received funding from the European Union’s Horizon 2020 research and innovation programme under grant agreement No 824087 – European Open Science Cloud in Life Sciences.
c Neurospin 7T received funding from the France-Life-Imaging project – grant 11-INBS-0006
d This work has been supported by the Leducq Foundation large equipment ERPT program, the NEUROVASC7T project, the Institut Carnot.
References
[1] Ravanfar, P., et al. (2021). Systematic review: quantitative susceptibility mapping (QSM) of brain iron profile in neurodegenerative diseases. Frontiers in neuroscience, 41.
[2] Wang, C., et al. (2022). Phenotypic and genetic associations of quantitative magnetic susceptibility in UK Biobank brain imaging. Nature Neuroscience, 1-14.
[3] Ruetten, P., et al. (2019). Introduction to quantitative susceptibility mapping and susceptibility weighted imaging. The British Journal of Radiology, 92(1101), 20181016.
[4] Wang, Y., et al. (2009, September). Magnetic source MRI: a new quantitative imaging of magnetic biomarkers. In 2009 Annual International Conference of the IEEE Engineering in Medicine and Biology Society (pp. 53-56). IEEE.
[5] Zachariou, V., Bauer, C. E., Powell, D. K., & Gold, B. T. (2022). Ironsmith: An automated pipeline for QSM-based data analyses. NeuroImage, 249, 118835.
[6] Liu, T., et al. (2011). Morphology enabled dipole inversion (MEDI) from a single‐angle acquisition: comparison with COSMOS in human brain imaging. Magnetic resonance in medicine, 66(3), 777-783.
[7] Haeger, A., et al. (2020). Imaging the aging brain: study design and baseline findings of the SENIOR cohort. Alzheimer's research & therapy, vol. 12(1), article 77, https://doi.org/10.1186/s13195-020-00642-1.
[8] Blaimer, M., et al. (2016). Comparison of phase constrained parallel MRI approaches: Analogies and differences. Magnetic resonance in medicine, 75(3), 1086-1099.
[9] de Rochefort, L., et al. (2009). Quantitative susceptibility mapping in vivo in the rat brain. Proc Intl Soc Magnet Reson Med, 17(c), 1134.
[10] Liu, T., et al. (2011). A novel background field removal method for MRI using projection onto dipole fields. NMR in Biomedicine, 24(9), 1129-1136.
Figures