3244
Reliability of brain volume measures of accelerated 3D T1-weighted images with deep learning-based reconstruction1AIRS Medical, Seoul, Korea, Republic of, 2Department of Radiology, Seoul National University Hospital, Seoul, Korea, Republic of
Synopsis
Keywords: Machine Learning/Artificial Intelligence, Brain
We validated the brain volumetric results with 3D T1 weighted images with accelerated scans reconstructed by FDA-cleared deep learning-based software (SwiftMR, AIRS Medical). Acceleration scans with three different acceleration levels were simulated using k-space undersampling, and the image quality and brain volume measures were evaluated. In addition, we acquired conventional and accelerated scans from each participant to evaluate the reliability between conventional and accelerated scans reconstructed by SwiftMR and inter-method reliability between different brain segmentation software. As a result, brain volume measures with accelerated scans with deep learning-based reconstruction were in good agreement with those of the corresponding conventional scan.Introduciton
Brain volume analysis based on 3D T1-weighted images is a completely noninvasive and widely used method to exclude other structural abnormalities and diagnose neurodegeneration. Therefore, several commercially available software were developed to reduce processing time and generalize their use in routine clinical practice [1]. Recently, several deep learning (DL)-based reconstruction algorithms have been introduced to accelerate 3D MR image acquisition. However, only a few studies evaluate the robustness of volumetry algorithms to DL-based 3D images [2,3]. In addition, inter-method reliability of different commercially available software for volume measurements has not been investigated using DL-based 3D images. In this study, we explored the compatibility of automated brain volume measures using the DL-based MR images by applying various acceleration times. Furthermore, we aimed to evaluate the inter-method reliability of DL-based accelerated 3D images using different software.Methods
[Data acquisition] This retrospective study included 90 subjects without visible focal lesions in the brain, comprising 42 consecutive subjects with conventional 3D T1-weighted MRI with MPRAGE sequence (Conv) for the simulation and 48 consecutive subjects with both Conv and accelerated 3D MPRAGE (Accel) for the validation study. Conv with k-space data was acquired as follows: TR/TE/TI = 1600~1740/2.8/900 ms, flip angle = 9°, voxel size = 1 x 1 x 1 mm3, phase resolution = 100%, GRAPPA factor = 2, and scan time = 180 ~ 214 s. The Accel protocol was determined by modifying the conventional protocol: GRAPPA factor = 3, phase resolution = 60%, and scan time = 100 ~ 119 s.[Software] For DL-based MR image reconstruction, DICOM-based FDA-cleared software (SwiftMR™, AIRS Medical) was utilized. The model was constructed as 2.5D U-net architecture and trained by 3D MP-RAGE brain images of around 1,000 participants acquired from 1.5T to 3T SIEMENS scanners. For volumetric MRI analysis, two clinically available software with different machine learning-based algorithms were utilized: NeuroQuant® (Cortechs.ai) and Deepbrain® (Vuno).
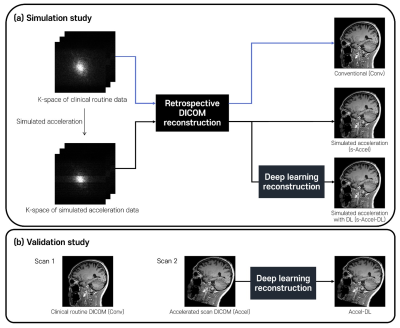
[Simulation study] To evaluate the measures on various acceleration times, k-space data from conventional scans were utilized by applying undersampling simulations and retrospective DICOM reconstruction (Figure 1a). Three types of images were generated from each k-space: conventional images (Conv), simulated-acceleration images (s-Accel), and simulated-acceleration images with DL-based reconstruction (s-Accel-DL). Note that 3 different acceleration times were simulated from 65% to 75% relative to full-sampled acquisition as follows:
Level 1: GRAPPA factor = 2, phase resolution = 60 %, simulated scan time = 128 ~ 152 s
Level 2: GRAPPA factor = 2, phase resolution = 50 %, simulated scan time = 109 ~ 130 s
Level 3: GRAPPA factor = 3, phase resolution = 60 %, simulated scan time = 91 ~ 109 s
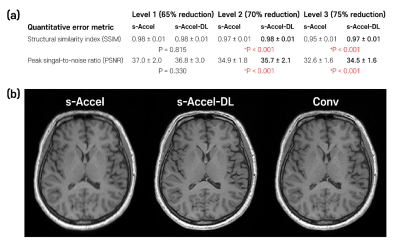
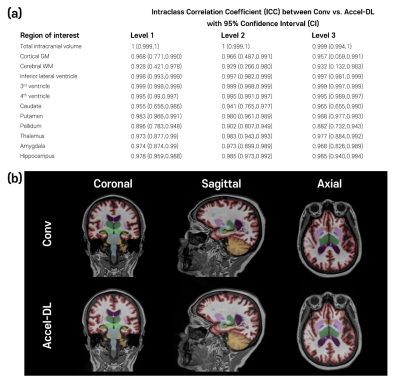
The acceleration level was determined based on a previous DL-based volumetry study [2]. The performance of s-Accel-DL was evaluated by measuring quantitative error metrics, structural similarity index (SSIM) and peak signal-to-noise ratio (PSNR). For statistical significance, a paired t-test was performed with Bonferroni correction. In addition, Conv and s-Accel-DL data were analyzed with NeuroQuant and Deepbrain, respectively. Intraclass correlation coefficients (ICCs) of regional brain volume measures were calculated between the Conv and s-Accel-DL. ICC is defined as poor (<0.5), moderate (0.50-0.75), good (0.75-0.90), and excellent (>0.9) [4].
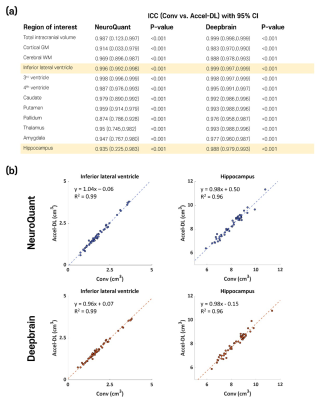
[Validation study] In the subjects with both Conv and Accel, accelerated scans with DL (Accel-DL) were constructed by applying the most adequate acceleration time determined on the simulation analyses (i.e., Acceleration level 3). To validate the brain volume measures from Accel-DL, ICCs were calculated to test inter-scan reliability between regional brain volume measures from Conv and Accel-DL. The linear regression analysis was performed between the estimated volume of Conv and Accel-DL in the two representative ROIs (i.e., inferior lateral ventricle and hippocampus). The inter-method reliability between NeuroQuant and Deepbrain was also calculated using Conv and Accel-DL, respectively.
Results
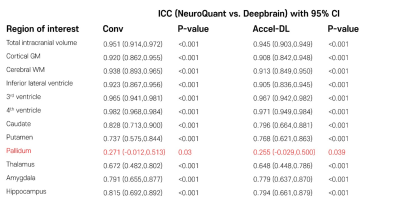
In the first simulation study, s-Accel-DL showed higher SSIM and PSNR than s-Accel as the acceleration level increased (in 75% reduction, SSIM/PSNR = 0.97/34.5 at s-Accel-DL and 0.95/32.6 at s-Accel, all P < 0.001, a paired t-test), revealing the better image quality (Figure 2). For the brain volume measurement with the simulated acceleration dataset, all ICC values between Conv and s-Accel-DL were good or excellent (> 0.88) in every acceleration level (Figure 3). In the second validation study, all ICC values between the volumetric results from Conv and Accel-DL were good or excellent (> 0.87) using both NeuroQuant and Deepbrain (Figure 4a). Particularly, Alzheimer’s disease-related regions such hippocampus, and inferior lateral ventricle [2,5] showed good agreement in linear regression analysis (R2 > 0.95, Figure 4b). Lastly, inter-method reliability between NeuroQuant and Deepbrain was moderate to excellent with both Conv and Accel-DL (ICC: 0.672-0.982 with Conv, 0.648-0.971 with Accel-DL, Figure 5) except the pallidum (ICC < 0.3).Discussion and Conclusion
DL-based reconstruction enhanced various acceleration times by up to 75% relative to full-sampled acquisition. Brain volume measures with Accel-DL images were in good to excellent agreement with those of Conv, regardless of the type of brain segmentation software. This finding supports the clinical feasibility of DL-accel images for the use of volumetric quantitative MRI in routine clinical practice.Acknowledgements
This work was supported by the Korea Medical Device Development Fund grant funded by the Korea government (the Ministry of Science and ICT, the Ministry of Trade, Industry and Energy, the Ministry of Health & Welfare, the Ministry of Food and Drug Safety) (Project Number: 9991006735 , RS-2020-KD000062)References
[1] Brewer, James B., et al. "Fully-automated quantification of regional brain volumes for improved detection of focal atrophy in Alzheimer disease." American Journal of Neuroradiology 30.3 (2009): 578-580.
[2] Bash, S., et al. "Deep learning enables 60% accelerated volumetric brain MRI while preserving quantitative performance: a prospective, multicenter, multireader trial." American Journal of Neuroradiology 42.12 (2021): 2130-2137.
[3] Lebel, R. Marc, et al. "Deep learning reconstruction enables accelerated acquisitions with consistent volumetric measurements." Joint Annual Meeting ISMRM-ESMRMB ISMRT 31st Annual Meeting (2022): 1060.
[4] Bartko, John J. "The intraclass correlation coefficient as a measure of reliability." Psychological reports 19.1 (1966): 3-11.
[5] Mofrad, Samaneh Abolpour, et al. "A predictive framework based on brain volume trajectories enabling early detection of Alzheimer's disease." Computerized Medical Imaging and Graphics 90 (2021): 101910.
Figures