3233
Conversion of legacy data of nonclinical trials using MRI into CDISC SEND: Toward Standardization of an Animal MR Imaging Data
Do-Wan Lee1, Young Chul Cho2, Mi-hyun Kim3,4, Yeon Ji Chae5, Su Jung Ham3, Yousun Ko1, Seongwon Na2, Youngbin Shin2, Nari Kim6, Dong‐Cheol Woo5,7, and Kyung Won Kim2
1Department of Radiology, Asan Medical Center, University of Ulsan College of Medicine, Seoul, Korea, Republic of, 2Biomedical Research Center, Asan Institute for Life Sciences, Asan Medical Center, Seoul, Korea, Republic of, 3Trialinformatics Inc., Seoul, Korea, Republic of, 4Department of Radiation Science & Technology, Jeonbuk National University, Jeonju, Korea, Republic of, 5Department of Convergence Medicine, Asan Medical Center, University of Ulsan College of Medicine, Seoul, Korea, Republic of, 6Department of Medical Science, Asan Medical Institute of Convergence Science and Technology, Asan Medical Center, University of Ulsan College of Medicine, Seoul, Korea, Republic of, 7Convergence Medicine Research Center, Asan Institute for Life Sciences, Asan Medical Center, Seoul, Korea, Republic of
1Department of Radiology, Asan Medical Center, University of Ulsan College of Medicine, Seoul, Korea, Republic of, 2Biomedical Research Center, Asan Institute for Life Sciences, Asan Medical Center, Seoul, Korea, Republic of, 3Trialinformatics Inc., Seoul, Korea, Republic of, 4Department of Radiation Science & Technology, Jeonbuk National University, Jeonju, Korea, Republic of, 5Department of Convergence Medicine, Asan Medical Center, University of Ulsan College of Medicine, Seoul, Korea, Republic of, 6Department of Medical Science, Asan Medical Institute of Convergence Science and Technology, Asan Medical Center, University of Ulsan College of Medicine, Seoul, Korea, Republic of, 7Convergence Medicine Research Center, Asan Institute for Life Sciences, Asan Medical Center, Seoul, Korea, Republic of
Synopsis
Keywords: Visualization, Visualization
In nonclinical animal research for new drug development, the US FDA mandates to manage data in accordance with clinical data interchange standards consortium (CDISC) standard for exchange of nonclinical data (SEND). Older nonclinical trial data, i.e., legacy nonclinical trial data, contain highly useful information but those data were created in various data formats by different researchers. The present study specifically describes how to implement the CDISC SEND data standards for the legacy nonclinical trial data with MRI. Standardization of animal MRI data with CDISC SEND enables us to utilize legacy data for next-generation drug development and new knowledge creation.INTRODUCTION
To date, magnetic resonance imaging (MRI) technology has made a great contribution not only to clinical trials, but also to various nonclinical trials for evaluation of new drug response assessment 1,2. Moreover, the use of MRI has been increasing in nonclinical trials for new drug development due to its high-resolution images with exquisite soft tissue contrast 3. However, there has been no standards in managing those experimental data. Thus, nonclinical trials with animal MRI have been accumulated with various data formats 2. To review these data with various data format, the United States Food and Drug Administration (U.S. FDA) requires significant time, cost, and human resources 4. To overcome these limitations, the clinical data interchange standards consortium (CDISC) established global industry standards to support the electronic data acquisition, submission, exchange and archiving of trial related documents and metadata for medical and biopharmaceutical product development 5. The U.S. FDA started requiring the submission of electronic data in accordance with the CDISC standard for trials beginning on or after December 17, 2016. Accordingly, many pharmaceutical companies have tried to convert their nonclinical data into the CDISC SEND standard 5. Therefore, animal MRI researchers should be familiar with CDISC SEND, especially in experiments for drug development. The present study specifically describes how to convert animal MRI experiment data in accordance with CDISC SEND standards.METHODS
A legacy nonclinical MRI experiment dataset from a study published in the Radiology journal was used for the CDISC SEND implementation 6. According to the SEND implementation guide version 3.1 (SENDIG v3.1), all test results, examinations, and observations for subjects in a nonclinical study are expressed in a series of SEND domains (Fig. 1). The SEND datasets are named to be consistent with the domain code (two-character identifier) and contain various unique variables. In addition, we generated domains for the ‘Trial Design’, ‘Demographics’, ‘Subject Elements’, ‘Exposure’, and ‘Disposition’ domains. ‘Findings Domains’, ‘Comments’ and ‘Relationships’, which are required by the CDISC SEND. After completing the data conversion, an expert in CDISC validated the converted data and data format.RESULTS
From the source data of our legacy nonclinical trial (Fig. 2a), all documents with study protocol, raw data including laboratory data and MRI finding, and every related finding (Fig. 2b, c, d) were recorded in a tabulated format according to the CDISC SENDIG v3.1. The MRI specific variables such as sequences and acquisition parameters were converted according to the SDTM for Medical Devices (SDTMIG-MD). The SEND datasets were converted to SAS-XPT format, and all terms used in tabulation were defined in the define-XML file 7. The SEND datasets in define-XML file were visualized to a website using the R Shiny application tool. Figure 3 shows the visualized demographic (DM) domain of the implemented SEND dataset on a website, which can facilitate efficient access by those involved in the planning, programming and validation phases of the conversion. Validation by an expert showed that all data were perfectly converted to CDISC SEND without any error.CONCLUSION
We successfully converted the legacy nonclinical trial with animal MRI into CDISC SEND data format and visualized them in a web. Implementation of the SEND format for animal MRI experiments is expected to increase in the field of drug development.Acknowledgements
This work was supported by the National Research Foundation of Korea (NRF) grant funded by the Korea Government (Ministry of Science and ICT, MSIT) (No. 2022R1C1C2008801).References
1. M Yamamoto, K Nagai, TI Saito. Implementation of Clinical Data Interchange Standards Consortium (CDISC) Study Data Tabulation Model (SDTM) in Investigator-Initiated Studies on Hematologic Malignancies. Available at SSRN 4233978. 2. Souza T, Kush R, Evans JP. Global clinical data interchange standards are here! Drug Discov. 2007;12(3-4);174-181. 3. Pavuluri K, Manoli I, Pass A, et al. Noninvasive monitoring of chronic kidney disease using pH and perfusion imaging. Sci Adv. 2019;5(8):eaaw8357. 4. Wood R, Guinter T. Evolution and Implementation of the CDISC Study Data Tabulation Model (SDTM). Pharmaceutical Programming. 2008;1(1):20-27. 5. Jeong S, Choi I, Kim S. International Standard in Electronic Clinical Trial. Kor J Clin Pharmacol Ther. 2007;15(1):20-27. 6. Lee DW, Heo H, Woo DC, et al. Amide Proton Transfer-weighted 7-T MRI Contrast of Myelination after Cuprizone Administration. Radiology. 2021 May;299(2):428-434. 7. Carfagna MA, Anderson J, Eley C, et al. Leveraging the Value of CDISC SEND Data Sets for Cross-Study Analysis: Incidence of Microscopic Findings in Control Animals. Chem Res Toxicol. 2021;34(2):483–494.Figures

Figure 1. Schematic diagrams indicate the defined domains based on the
standard for exchange of nonclinical data (SEND) implementation guide version
3.1. Blue (*) and red (**) font reflects the relevant domains of in-life- and
post-mortem data, respectively.
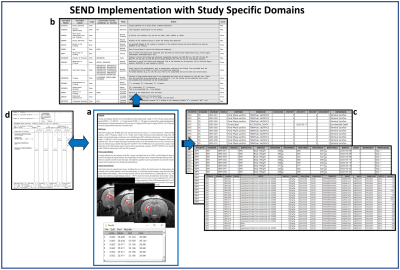
Figure 2. A schematic representation of the SEND implementation using
the documents of laboratory data and protocol design. SEND, standard for
exchange of nonclinical data
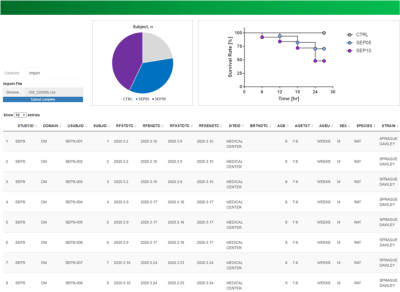
Figure 3. Screenshots shows R Shiny application user interface with
demographic (DM) summary data table displayed.
DOI: https://doi.org/10.58530/2023/3233