3222
Spatial habitats features derived from multiparametric MR imaging predict long versus short-term survival in gliomas: Preliminary findings1Department of Radiology, The First Affiliated Hospital, Sun Yat-sen University, Guangzhou, China, 2MR Scientific Marketing, Siemens Healthineers Ltd., Guangzhou, China, 3Shanghai Key Laboratory of Magneitc Resonance, East China Normal University, Shanghai, China
Synopsis
Keywords: Tumors, Brain, high-grade gliomas, tumor habitats, habitat imaging
We developed spatial habitats based on multiparametric MRI and evaluated associations between features in these habitats and survival time in patients with high-grade gliomas. The voxels in MR images were grouped into 2 clusters using the K-means clustering algorithm of in-house software nnFAE (V.0.0.10). Structural MRI habitats were defined on CE-T1WI and T2-Flair images, and physiologic MRI habitats were defined on MK derived from DKI and Ktrans derived from DCE imaging. Results showed physiologic habitats weighed more than structural ones, and suggested low vascular-permeability-and-tissue-complexity habitats may play an important role in distinguishing long- and short-term survival of high-grade gliomas patients.Introduction
High-grade (including grade of WHO 4) glioma (HGG) is the most aggressive intracranial tumours, easy to relapse and with poor prognosis [1]. Their median survival is about 12-15 months. So it has important clinical significance to predict prognosis of HGG. Significant heterogeneity of HGG is associated with poor outcomes and multiparametric MRI has been confirmed that can reflect HGG heterogeneity. Radiomics can noninvasively quantified heterogeneity by capturing subvisual cues of morphologic diversity (eg, roughness and edges) in patients [2-4]. However, features extraction often occurs in the whole tumor region using radiomics, so that it is not sufficient to clearly express partially spatial heterogeneity. And then, the tumor habitat imaging analysis come into being [5]. It can divide the tumor into several subregions with the same or similar heterogeneity based on different pathophysiological characteristic which ideally reveals the spatial heterogeneity within the tumor and ensures the homogeneity within the subregions to the greatest extent. It has been reported that habitat features based on multiparametric contrast-enhanced T1-weighted imaging (CE-T1WI), T2-fluid-attenuated inversion recovery (T2-Flair), apparent diffusion coefficient (ADC), and cerebral blood volume (CBV) images can better predict the overall survival (OS) in high-grade glioma [6-11].Purpose
We hypothesized that features of tumor habitats derived from structural and physiologic MRI such as CE-T1WI, T2-Flair, mean kurtosis(MK), and volume transfer constant (Ktrans) could characterize the spatial complexity of the heterogeneity of HGG and can be used to predict the survival time of patients. The purpose of this study was to investigate the associations between image-based spatial habitats characteristics and overall survival (OS) in patients with HGG.Methods
Seven participants were preliminarily included into this study and were divided into LTSs (OS ≥ 600 days, n=4) and STSs (OS < 600 days, n=3). They were all diagnosed as high-grade diffuse gliomas of WHO grade 4 according to the latest classification edition. Ktrans parameter maps derived from DCE imagings and MK parameter maps derived from DKI imagings were obtained. For each patient, all images were coregistered with the T1-spatial mapping images and ROIs of the CE portions were manually drawed. The voxels in the ROIs were then aggregated into clusters using a K-means clustering module in our in-house software nnFAE (V.0.0.10). Features were extracted by FAE software (v.0.5.3). In this work, we defined structural tumor habitats derived from Ktrans and MK feature maps and physiologic tumor habitats derived from CE-T1W1 and T2-Flair images. Finally, Mann-Whitney, the Receiver Operating Characteristics (ROC) curves and Kaplan–Meier survival analysis were carried out.Results
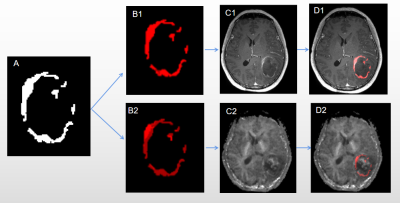
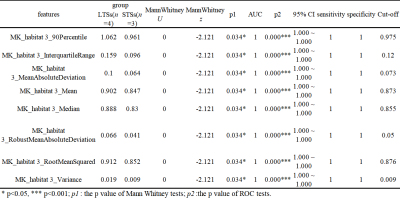
As shown in Fig. 1, two clusters were applied for structural and physiologic MRI habitats. In Fig.1 E1, orange (habitat 1) and blue areas (habitat 2) on the volume fractions of the structural MRI represent low-enhancing and high-enhancing tissue habitats, respectively. And in Fig.1 E2, orange (habitat 3) and blue areas (habitat 4) on the volume fractions of the physiologic MRI represent low or high vascular-permeability-and-tissue-complexity, respectively. We then applied K-means clustering onto original MR images (Fig.2 and 3) and the four habitats were given different colors.Many MR features including Percent, Volume, Variance, Skewness and so on of different MR sequence from different habitats and were extracted. Many features derived from physiologic MRI habitat were marginally statistically significant between LTSs and STSs, while only one feature derived from structural MRI habitats namely kurtosis of CE-T1 from habitat 1 were marginally statistically significant. And these features all performed high-sensitivity (even 1) and high-specificity ( even 1) in distinguishing LTSs and STSs. More MR feature devired from MK images on habitat 3 performed well to distingushing LTSs (Table 1).
Discussion
This was a preliminary feasibility study of tumor habitat analysis, using DCE and DKI imaging which is the first attempt to our knowledge, to predicting long versus short-term survival in gliomas. And the in-house software nnFAE was independently researched and developed by the research team of our co-authors and it is also the first attempt for this software about studying glomas.The results suggest that physiologic MRI habitats performed better than structural MRI habitats in differentiating LTSs and STSs. MK features mainly played an important role rathan than Ktrans features. And for LTSs, MK features derived from habitat 3 which represented the low vascular-permeability-and-tissue-complexity of tumors weighed much.
The MK values were relatively smaller in habitat 3 than those in habitat 4. So tumors portions in habitat 3 may have lower cell density, less abundant vascular proliferation, and less obvious cellular atypia, and thus may be correlated with good prognosis. ·
In further research, more participators, more clusters and in-depth description of statistics should be applied and chosen for obtaining the best physiologic MRI habitats relevant with prognosis.
Conclusion
Low vascular-permeability-and-tissue-complexity habitats derived from multi-parametric physiologic MRIs of Ktrans and MK images may be useful predictors of clinical outcomes in patients with gliomas.Acknowledgements
This study was supported by the Guangdong Basic and Applied Basic Research Foundation, China (No.2020A1515011436, 2021A1515012279, 2022A1515011264) and the National Natural Science Foundation (NSFC 82172015).References
[1] The Medical Administration of the National Health Commission, the Glioma Professional Committee of China Anti-Cancer Association, and the Glioma Professional Committee of Chinese Physicians Association. Guidelines for diagnosis and treatment of brain Glioma (2022 Edition) [J]. Chinese Journal of Neurosurgery, 2022, 38 (08): 757-777.
[2] Verma, R. et al. Tumor Habitat–derived Radiomic Features at Pretreatment MRI That Are Prognostic for Progression-free Survival in Glioblastoma Are Associated with Key Morphologic Attributes at Histopathologic Examination: A Feasibility Study. Radiol. Artif. Intell. 2, e190168 (2020).
[3] Prasanna, P., Patel, J., Partovi, S., Madabhushi, A. & Tiwari, P. Radiomic features from the peritumoral brain parenchyma on treatment-naïve multi-parametric MR imaging predict long versus short-term survival in glioblastoma multiforme: Preliminary findings. Eur. Radiol. 27, 4188–4197 (2017).
[4] Mcgranahan N, Swanton C. Clonal heterogeneity and tumor evolution: past, present, and the future. Cell, 2017, 168: 613 -628. DOI:10.1016/j.cell.2017.01.018.
[5] Sala, E. et al. Unravelling tumour heterogeneity using next-generation imaging: radiomics, radiogenomics, and habitat imaging. Clin. Radiol. 72, 3–10 (2017).
[6] Chang YC, Ackerstaff E, Tschudi Y, et al. Delineation of tumor habitats based on dynamic contrast enhanced MRI. Sci Rep,2017, 7: 9746. DOI:10.1038/s41598-017-09932-5.
[7] Stringfield, O. et al. Multiparameter MRI Predictors of Long-Term Survival in Glioblastoma Multiforme. Tomography 5, 135–144 (2019).
[8] Lee, D. H. et al. Tumor habitat analysis by magnetic resonance imaging distinguishes tumor progression from radiation necrosis in brain metastases after stereotactic radiosurgery. Eur. Radiol. 32, 497–507 (2022).
[9] Xu, X., Samaras, D. & Prasanna, P. Radiologically Defined Tumor-habitat Adjacency as a Prognostic Biomarker in Glioblastoma. in 2021 43rd Annual International Conference of the IEEE Engineering in Medicine & Biology Society (EMBC) 3998–4001 (IEEE, 2021). doi:10.1109/EMBC46164.2021.9629779.
[10] Park, J. E. et al. Spatiotemporal Heterogeneity in Multiparametric Physiologic MRI Is Associated with Patient Outcomes in IDH-Wildtype Glioblastoma. Clin. Cancer Res. 27, 237–245 (2021).
[11] Juan-Albarracin J, Fuster -Garcia E, Perez -Girbes A, et al. Glioblastoma: vascular habitats detected at preoperative dynamic susceptibility-weighted contrast-enhanced perfusion MR imagingpredict survival[J]. Radiology, 2018, 287: 944 -954. DOI:10.1148/radiol.2017170845.
Figures
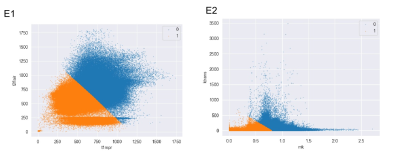
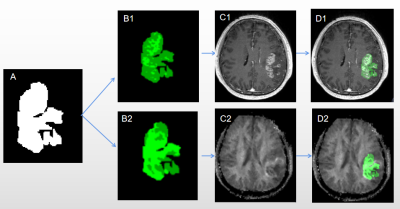
Figure 2. A representative example of STSs from a 55-year-old patient. A ROIs drawn on CE-T1WI portions; B1 Structural MRI habitats;B2 Physiologic MRI habitats; C1 CE-T1WI; C2 MK image; K-means clustering is applied to CE-T1-weighted (D1); K-means clustering is applied to MK images (D2)