3137
Deep learning-based image quality and spatial resolution improvement for Diffusion Weighted Imaging in liver1Philips Japan, Tokyo, Japan, 2Tokyo Metropolitan Police Hospital, Nakano, Japan, 3Philips Healthcare, Best, Netherlands
Synopsis
Keywords: Liver, Diffusion/other diffusion imaging techniques
Diffusion-weighted imaging (DWI) in liver plays a significant role for lesion characterization and staging of fibrosis. Single-shot echo-planar imaging (ssh-EPI) readout is typically used; however, spatial resolution of ssh-EPI-DWI is limited by acquisition time. In this study, we investigated the use of prototype AI-based reconstruction technique (SmartSpeed Precise Image) to improve the image quality of liver ssh-EPI-DWI images. The image quality was compared between conventional Compressed-SENSE (C-SENSE), SmartSpeed AI, and SmartSpeed Precise Image. Volunteer data demonstrated a significant improvement of sharpness in DWI images and ADC map, and reduction of ringing artifact compared with C-SENSE and SmartSpeed AI reconstruction.
Introduction
In recent years there has been growing interest in abdominal diffusion-weighted imaging (DWI). Abdominal DWI is reported to be useful for liver lesion detection and characterization1–4, and evaluation of fibrosis and inflammation5–7. Single-shot echo-planar imaging (ssh-EPI) readout is typically applied for liver DWI to mitigate motion. However, spatial resolution of ssh-EPI DWI is inherently limited by acquisition time, even with the use of scan acceleration techniques such as parallel imaging8 or compressed sensing9,10. Although apparent spatial resolution is improved by using zero-filling interpolation (ZIP), the improvement of the actual spatial resolution is limited.In the last years there has been a growing interest in using Artificial Intelligence (AI) to improve the spatial resolution of medical images11. In the context of DWI, this so-called “SuperResolution AI” demonstrated enhanced delineation of breast cancer and the boundary of surrounding structure12. However, there are very few studies applying AI for liver DWI.
In this study, two types of AI were used to first remove the noise and undersampling artifacts from the image and subsequently to improve the sharpness of the image. It is hypothesized that the image quality of liver DWI and ADC map can be significantly improved by using the proposed AI-based reconstruction technique. The purpose of this study was to acquire high-resolution and high-SNR liver DWI images using the proposed AI-based reconstruction, and to compare the image quality with conventional Compressed-SENSE (C-SENSE) and SmartSpeed AI reconstructions.
Methods
The study was approved by the local IRB, and written informed consent was obtained from all subjects. A total of 5 volunteers were examined on a 3.0T whole-body clinical system (Ingenia Elition X, Philips Healthcare) using a 32-channel phased-array coil. Multi-slice 2D ssh-EPI-DWI sequence was acquired in the axial plane.Images have been reconstructed using vendor provided prototype software (Philips SmartSpeed Precise Image). This AI-based reconstruction technique consists of a series of convolutional neural networks (CNNs): Adaptive-CS-Net13 allows to reconstruct images acquired with Compressed SENSE based variable density undersampling patterns. This CNN is applied during the coil combination, removing the noise and related undersampling artifacts from the images in order to obtain good image quality from accelerated acquisitions14. Subsequently, Precise Image Net is an AI-model applied to remove ringing artefacts and to replace the traditional zero-filling strategy to increase the matrix size and therewith the sharpness of the images; these type of networks are known as SuperResolution networks11,15. This network is trained on pairs of low- and high-resolution data with k-space crops to induce ringing.
Data consistency checks are implemented to match the resulting k-space with the measured k-space data. The full reconstruction pipeline generates images with improved SNR and sharpness, higher matrix size and reduced ringing artefacts and can be applied to all 2D cartesian acquisitions. The noise reduction can be controlled using a parameter to the users’ preference.
The image quality was compared between three acceleration methods: C-SENSE, SmartSpeed AI, and SmartSpeed Precise Image. Scans were respiratory triggered with a trigger delay of 300ms. Following parameters were common to all examinations: FOV=360x360mm2, slice thickness=7mm, 25 slices, acquisition resolution=3.5×3.5mm2, reconstruction resolution=1.6×1.6 and 0.83×0.83mm2, b-value=0 and 800 s/mm2, TR=1306ms, TE=49ms, acceleration factor=2.5, number of signals averaged=5, and the scan time was 1min48sec.
To quantitatively evaluate the influence of the reconstruction techniques on DWI signal intensities and ADC values, the entire liver of one subject was manually segmented using 3D Slicer16,17. The segmentations were then overlayed to DWI images and ADC maps, and histogram analysis was conducted.
Results and Discussions
Figure 1 shows comparison of C-SENSE, SmartSpeed AI, and SmartSpeed Precise Image, all reconstructed with 1.6x1.6mm2 spatial resolution. Visually, SmartSpeed Precise Image showed improved sharpness in both b=0 and 800 s/mm2 images as well as ADC map. Figures 2 and 3 show comparisons of C-SENSE, SmartSpeed AI, and SmartSpeed Precise Image, all reconstructed with 0.83x0.83mm2 spatial resolution. The higher reconstruction resolution allowed SmartSpeed Precise Image to delineate vascular pattern even more clearly at b=0 s/mm2 and enhanced the edges at b=800 s/mm2. This resulted in sharper ADC map compared with C-SENSE and SmartSpeed AI, which used ZIP to improve the resolution. Figure 4 shows the enlarged view of b=0 s/mm2 images and ADC maps of C-SENSE, SmartSpeed AI, and SmartSpeed Precise Image, all reconstructed with 0.83x0.83mm2 spatial resolution. SmartSpeed Precise Image showed significantly improved visualization of vessels in both b=0 s/mm2 image and ADC map, which may enable evaluating smaller vascular patterns that were previously unobservable. Severe ringing artifacts caused by stomach contents and cerebrospinal fluid were significantly reduced by SmartSpeed Precise Image (Figures 2 and 4, yellow arrows). Figure 5 shows the histogram of the DWI pixel values and ADC values of the liver. In this subject, the mean pixel values for b=0 s/mm2 were 294.4, 294.2, 286.8 for C-SENSE, SmartSpeed AI, and SmartSpeed Precise Image, respectively, and for b=800 s/mm2 were 80.2, 83.0, 83.6, and the mean ADC values were 1.66, 1.61, 1.54 x10-3 mm2/s.Conclusion
High-resolution and high-SNR DWI images for the liver were reconstructed using vendor provided prototype AI-based reconstruction software. Volunteer data demonstrated significantly improved sharpness of DWI images and ADC maps compared with conventional techniques, which resulted in the improved visualization of small vascular patterns.Acknowledgements
No acknowledgement found.References
1. Coenegrachts K, Delanote J, Ter Beek L, et al. Improved focal liver lesion detection: Comparison of single-shot diffusion-weighted echoplanar and single-shot T2 weighted turbo spin echo techniques. Br J Radiol. 2007;80(955):524-531. doi:10.1259/bjr/33156643
2. Bruegel M, Holzapfel K, Gaa J, et al. Characterization of focal liver lesions by ADC measurements using a respiratory triggered diffusion-weighted single-shot echo-planar MR imaging technique. Eur Radiol. 2008;18(3):477-485. doi:10.1007/s00330-007-0785-9
3. Vandecaveye V, De Keyzer F, Verslype C, et al. Diffusion-weighted MRI provides additional value to conventional dynamic contrast-enhanced MRI for detection of hepatocellular carcinoma. Eur Radiol. 2009;19(10):2456-2466. doi:10.1007/s00330-009-1431-5
4. Galea N, Cantisani V, Taouli B. Liver lesion detection and characterization: Role of diffusion-weighted imaging. J Magn Reson Imaging. 2013;37(6):1260-1276. doi:10.1002/jmri.23947
5. Taouli B, Chouli M, Martin AJ, Qayyum A, Coakley F V., Vilgrain V. Chronic hepatitis: Role of diffusion-weighted imaging and diffusion tensor imaging for the diagnosis of liver fibrosis and inflammation. J Magn Reson Imaging. 2008;28(1):89-95. doi:10.1002/jmri.21227
6. Bakan AA, Inci E, Bakan S, Gokturk S, Cimilli T. Utility of diffusion-weighted imaging in the evaluation of liver fibrosis. Eur Radiol. 2012;22(3):682-687. doi:10.1007/s00330-011-2295-z
7. Ichikawa S, Motosugi U, Morisaka H, et al. MRI-based staging of hepatic fibrosis: Comparison of intravoxel incoherent motion diffusion-weighted imaging with magnetic resonance elastography. J Magn Reson Imaging. 2015;42(1):204-210. doi:10.1002/jmri.24760
8. Pruessmann KP, Weiger M, Börnert P, Boesiger P. Advances in sensitivity encoding with arbitrary k-space trajectories. Magn Reson Med. 2001;46(4):638-651. doi:10.1002/mrm.1241
9. Lustig M, Donoho D, Pauly JM. Sparse MRI: The application of compressed sensing for rapid MR imaging. Magn Reson Med. 2007;58(6):1182-1195. doi:10.1002/mrm.21391
10. Chen N kuei, Guidon A, Chang HC, Song AW. A robust multi-shot scan strategy for high-resolution diffusion weighted MRI enabled by multiplexed sensitivity-encoding (MUSE). Neuroimage. 2013;72(1):41-47. doi:10.1016/j.neuroimage.2013.01.038
11. Li Y, Sixou B, Peyrin F. A Review of the Deep Learning Methods for Medical Images Super Resolution Problems. Irbm. 2021;42(2):120-133. doi:10.1016/j.irbm.2020.08.004
12. Fan M, Liu Z, Xu M, et al. Generative adversarial network-based super-resolution of diffusion-weighted imaging: Application to tumour radiomics in breast cancer. NMR Biomed. 2020;33(8):1-12. doi:10.1002/nbm.4345
13. Pezzotti N, de Weerdt E, Yousefi S, et al. Adaptive-CS-Net: FastMRI with Adaptive Intelligence. arxiv. 2019;(NeurIPS). http://arxiv.org/abs/1912.12259
14. Peeters H, Chung H, Valvano G, et al. Philips SmartSpeed No compromise and robustness.
15. Chaudhari AS, Fang Z, Kogan F, et al. Super-resolution musculoskeletal MRI using deep learning. Magn Reson Med. 2018;80(5):2139-2154. doi:10.1002/mrm.27178
16. Kikinis R, Pieper SD, Vosburgh KG. 3D Slicer: A Platform for Subject-Specific Image Analysis, Visualization, and Clinical Support. In: Intraoperative Imaging and Image-Guided Therapy. Springer New York; 2014:277-289. doi:10.1007/978-1-4614-7657-3_19
17. 3D Slicer image computing platform. https://www.slicer.org/
Figures
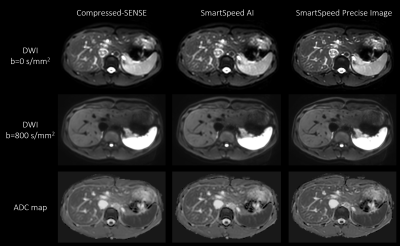
Figure 1. b = 0 (upper row), b = 800 s/mm2 (middle row) images, and ADC maps (lower row) of the liver in a healthy volunteer obtained with EPI-DWI, for C-SENSE (left), SmartSpeed AI (middle), and SmartSpeed Precise Image (right). All the images were reconstructed with 1.6x1.6 mm2 resolution.
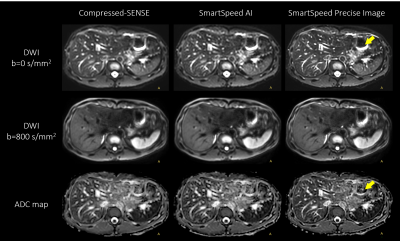
Figure 2. b = 0 (upper row), b = 800 s/mm2 (middle row) images, and ADC maps (lower row) of the liver in a healthy volunteer obtained with EPI-DWI, for C-SENSE (left), SmartSpeed AI (middle), and SmartSpeed Precise Image (right). All the images were reconstructed with 0.83x0.83 mm2 resolution. Arrows indicate the reduced ringing artifact in SmartSpeed Precise Image.
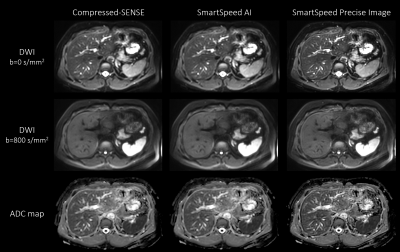
Figure 3. b = 0 (upper row), b = 800 s/mm2 (middle row) images, and ADC maps (lower row) of the liver in a healthy volunteer obtained with EPI-DWI, for C-SENSE (left), SmartSpeed AI (middle), and SmartSpeed Precise Image (right). All the images were reconstructed with 0.83x0.83 mm2 resolution.
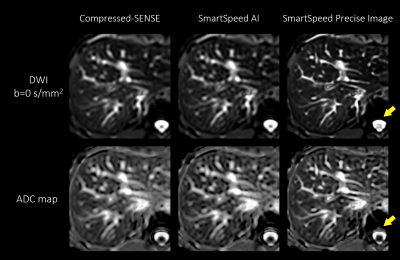
Figure 4. Enlarged view of b = 0 s/mm2 (upper row) images and ADC maps (lower row) of the liver in a healthy volunteer, for C-SENSE (left), SmartSpeed AI (middle), and SmartSpeed Precise Image (right). All the images were reconstructed with 0.83x0.83 mm2 resolution. Arrows indicate the reduced ringing artifact in SmartSpeed Precise Image.
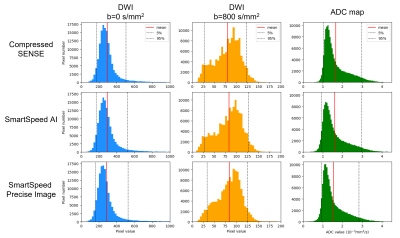
Figure 5. DWI pixel values and ADC values extracted from the entire liver are displayed in histogram for C-SENSE (top row), SmartSpeed AI (middle row), and SmartSpeed Precise Image (lower row). Black lines show 5 and 95 percentiles. Red lines show the average of the histogram.