3127
Improved pulmonary oxygen-enhanced MRI parameter precision enabled by hierarchical Bayesian inference1Bioxydyn Ltd, Manchester, United Kingdom, 2MCMR, Manchester University NHS Foundation Trust, Manchester, United Kingdom, 3National Heart & Lung Institute, Imperial College London, London, United Kingdom, 4Royal Brompton Hospital, Guy's & St Thomas’ Trust, London, United Kingdom, 5Centre for Medical Image Computing, Department of Medical Physics and Biomedical Engineering, University College London, London, United Kingdom
Synopsis
Keywords: Data Analysis, Lung, Bayes
Lung oxygen-enhanced MRI can provide regional information relating to lung function, but voxel-wise parameter estimation is hampered by low SNR. Here we present a hierarchical Bayesian approach to voxel-wise parameter estimation implemented in R and the probabilistic programming language Stan.
In both simulations and in OE-MRI data acquired in patients with cystic fibrosis, the Bayesian approach results in substantially less noisy parameter maps compared to conventional non-linear least squares estimation.
Introduction
Pulmonary oxygen-enhanced MRI (OE-MRI) uses inhaled oxygen as contrast agent to provide information relating to lung function. T2*-based methods1 are sensitive to changes in alveolar oxygen concentration and may provide a more specific measure of ventilation than T1-based methods2 but regional voxelwise analysis using non-linear least-squares (NLS) fitting is hampered by low SNR. Hierarchical Bayesian fitting techniques have been shown to improve voxelwise parameter estimates derived from noisy intravoxel incoherent motion (IVIM) data in the liver3 and the placenta4. Bayesian inference techniques use Markov Chain Monte Carlo (MCMC) methods to sample from a posterior distribution using methods such as Gibbs3,4 sampling or Hamiltonian Monte Carlo (HMC)5. The No-U-Turn Sampler5 (NUTS) is an extension of HMC which has been packaged into the probabilistic programming language Stan6. Here we apply a Bayesian approach to the estimation of regional changes in R2* using the R package brms7 which provides an interface to Stan and compare this with a conventional voxelwise NLS parameter estimation.Methods
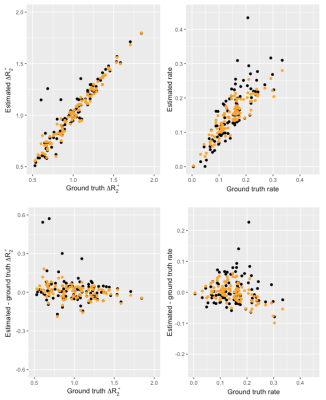
Dynamic OE-MRI simulations were performed to test the hierarchical Bayes implementation and compare with NLS with a known ground truth. 100 parameter pairs (∆R2* = R2*(oxygen) - R2*(air) and wash-in rate (α)) were simulated by random draws from a multivariate normal distribution, including a correlation between ∆R2* and α. Uptake curves were then generated using the equation ∆R2*dynamic(t) = ∆R2* (1 - exp(-t* α)), where t is time, with the addition of gaussian noise. Bayesian fitting was performed in R version 4.2.1 using the brms package. Normally distributed priors were specified for ∆R2* and α, with exponential priors for standard deviations and an LKJ8 prior for the correlation matrix. Three chains were run in parallel each with 2000 iterations in total, including 1000 for warm up. NLS fitting was performed using the R function nls().The patient dynamic OE-MRI data set consisted of 39 subjects with cystic fibrosis, scanned using a 1.5T Siemens Aera MRI scanner. Subjects were fitted with a non-rebreathing facemask (Intersurgical EcoliteTM, Intersurgical) and medical gases were supplied at 15l/min. Dual-echo multi-slice gradient-echo (TE=0.98,2.0ms, TR=16ms, FA=5deg, FOV=450x450mm, 5x10mm slices, 360 dynamics, temporal resolution 1.5s) were acquired dynamically and the medical gas supply was switched from air to 100% oxygen after 1.5min and back to air at 5min. Images were registered using ANTs9 and voxelwise dynamic R2* was calculated from the two echo time images. The voxelwise R2* prior to gas switching was averaged to obtain R2*(air) discarding the first 10 images of the dynamic series to ensure steady state had been achieved.
Prior to voxelwise analysis, the R2* curves were averages across all voxels in a lung slice and fitted using NLS. The resulting parameter estimates were used to provide the mean value for the priors for the Bayes fitting as well as the starting parameters for the voxelwise NLS fitting. All subjects also had pulmonary function testing (PFT) to obtain FEV1%, FVC% from spirometry and the lung clearance index (LCI2.5) from multiple breath washout. Here we present exemplar results for two subjects (subject 1: age=15, LCI2.5=9.3; subject 2: age=19, LCI2.5=11.4). Both subjects had a normal FEV1%.
Results
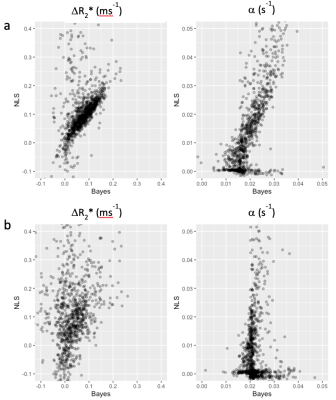
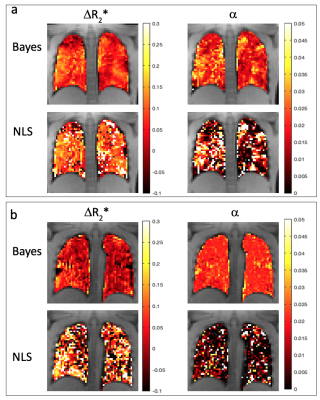
Figure 1 shows the comparison between Bayes and NLS fitting for the simulated data. The Bayesian fitting produced estimates that were on average closer to the ground truth and with fewer outliers. Figure 2 shows the calculated dynamic R2* averaged across a posterior slice in the lung for the two example subjects. A smaller change in R2* on oxygen breathing was observed in subject 2 compared to subject 1. A comparison between voxelwise estimates of ∆R2*and wash-in rate α for the two fitting methods is presented in figure 3 with the corresponding parameter maps in figure 4. The NLS fitting resulted in a much broader range of estimates for both parameter values than the Bayesian fitting and visually much more noisy parameter maps. Spatially varying parameter estimates are apparent in both patient datasets when using the Bayes approach that are obscured by noise when using NLS.Discussion
Bayesian methods are increasing in popularity for fitting of multi-voxel data. Using a hierarchical approach, the individual voxel and summary statistics are jointly modelled leading to more robust parameter estimation. Resulting parametric maps appear less noisy without the need to apply spatial smoothing to the data, so potentially preserving spatial features. As the priors for the Bayesian fitting were based on an initial ROI fit of the data, the Bayesian parameter estimates tend towards the ROI mean in very low SNR voxels, where the voxelwise estimates tend to be more dominated by the prior. This may explain the more homogeneous appearance of the wash-in rate parameter map in subject 2 compared to subject 1. The NLS fitting results in a large number of outliers and fit failures whereas the Bayes fitting results in parameter estimates closer to the ROI estimates.Conclusion
Bayesian parameter estimation using hierarchical non-linear modelling is straightforward to implement in R using the brms interface to Stan and may enable robust parametric mapping for noisy imaging data sets such as those obtained in lung OE-MRI. This robustness comes at the expense of substantially longer processing times (several hours vs seconds).Acknowledgements
Cystic Fibrosis Foundation, grant number 0208A120References
1. Pracht ED, Arnold JF, Wang T, Jakob PM. Oxygen-enhanced proton imaging of the human lung using T2. Magn Reson Med. 2005 May;53(5):1193-6.
2. Edelman RR, Hatabu H, Tadamura E, Li W, Prasad PV. Noninvasive assessment of regional ventilation in the human lung using oxygen-enhanced magnetic resonance imaging. Nat Med. 1996 Nov;2(11):1236-9.
3. Orton MR, Collins DJ, Koh DM, Leach MO. Improved intravoxel incoherent motion analysis of diffusion weighted imaging by data driven Bayesian modeling. Magn Reson Med. 2014 Jan;71(1):411-20
4. Flouri D, Owen D, Aughwane R, Mufti N, Maksym K, Sokolska M, Kendall G, Bainbridge A, Atkinson D, Vercauteren T, Ourselin S, David AL, Melbourne A. Improved fetal blood oxygenation and placental estimated measurements of diffusion-weighted MRI using data-driven Bayesian modeling. Magn Reson Med. 2020 Jun;83(6):2160-2172
5. Hoffman MD, Gelman A. The No-U-Turn sampler: adaptively setting path lengths in Hamiltonian Monte Carlo. J Mach Learn Res. 2014;15:1593–623.
6. Carpenter B, Gelman A, Hoffman MD, Lee D, Goodrich B, Betancourt M, et al. Stan: a probabilistic programming language. J Stat Softw. 2017;76:1–32.
7. Bürkner, P.-C. brms: An R Package for Bayesian Multilevel Models Using Stan. Journal of Statistical Software. 2017;80(1), 1–28.
8. Daniel Lewandowski, Dorota Kurowicka, Harry Joe. "Generating random correlation matrices based on vines and extended onion method." Journal of Multivariate Analysis. 2009;100(9), 1989-2001
9. Avants BB, Tustison NJ, Stauffer M, Song G, Wu B, Gee JC. The Insight ToolKit image registration framework. Front Neuroinform. 2014 Apr 28;8:44.
Figures
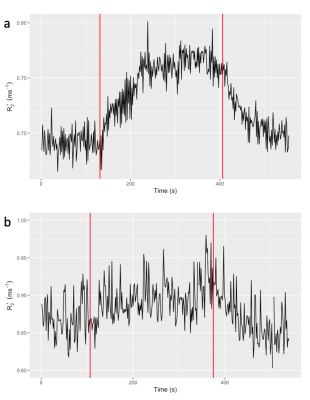
Mean lung R2* over the dynamic series for a) subject 1 and b) subject 2. Red lines indicate the gas switch points.