3122
A robust image analysis pipeline for high fidelity kurtosis and tensor fitting of 1.2mm isotropic infant brain diffusion MRI1Department of Radiology, Children's Hospital of Philadelphia, Philadelphia, PA, United States, 2Department of Bioengineering, University of Pennsylvania, Philadelphia, PA, United States, 3Department of Radiology, University of Pennsylvania, Philadelphia, PA, United States
Synopsis
Keywords: Data Processing, Pediatric, Neuro
High-resolution infant diffusion MRI (dMRI) data poses specific challenges such as severe motion artifacts, long acquisition time for multi-shell data, and low signal-to-noise (SNR). We present a robust image analysis pipeline for high fidelity kurtosis and tensor fitting of 1.2mm isotropic infant brain dMRI that allows us to efficiently analyze in-vivo dMRI data despite the considerable technical challenges specific to infant imaging. State-of-the-art preprocessing including slice-to-volume motion correction and susceptibility-by-movement correction is combined with advanced self-supervised learning-based denoising to produce high fidelity diffusion tensor and diffusion kurtosis fitting.Introduction
Diffusion magnetic resonance imaging (dMRI) on infant brains allows for non-invasively probing brain microstructure during the most dynamic period in brain development1. However, high-resolution infant dMRI data poses technical challenges such as low signal-to-noise ratio (SNR) and severe motion artifacts including within-volume motion and susceptibility-by-movement artifacts particularly due to long acquisition times for multi-shell data2-4. Smaller voxel sizes needed for imaging infant brain inherently leads to low SNR2. We present a robust image analysis pipeline for high fidelity kurtosis and tensor fitting that allows us to efficiently analyze in-vivo dMRI data at 1.2mm resolution despite the considerable technical challenges specific to infant imaging.Methods
High resolution 1.2mm Acquisition A highly optimized dMRI sequence (Multiband factor of 4, anterior-posterior (AP) and posterior-anterior (PA) phase encoding directions, 15 b0s, b values of 1000-2500s/mm2 , 64 AP and 96 PA dMRI volumes, 1.2mm isotropic resolution, 100 slices, TR/TE of 4750/113ms) developed for scanning infants during natural sleep has been implemented on a clinical 3T Siemens Prisma scanner. The acquisition has been applied to 100+ typically developing infants aged 0-25months.Comprehensive susceptibility, eddy current, and motion artifact correction
Correction of B0 susceptibility-induced artifacts
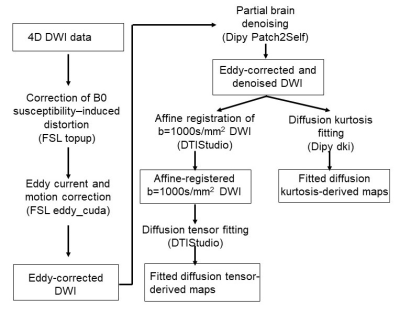
Fig. 1 shows the robust image analysis pipeline. Diffusion weighted images (DWI) first undergo correction of B0 susceptibility-induced distortion using two b0 volumes with opposite phase encoding (AP, PA). Field maps are estimated from the two b0 volumes using FSL’s topup function and applied to all diffusion volumes.
Systematic joint correction of eddy-current and motion artifacts
Joint eddy current and motion correction is carried out by a comprehensive framework that systematically accounts for various motion artifacts based on a Gaussian process data predictor. The framework simultaneously estimates subject motion and eddy currents5,6. Using the estimated distortion fields, the framework detects and replaces outlier slices with signal dropout7. Slice-to-volume motions (Fig. 2a) is accounted for by modelling movement as a piecewise continuous function over time8, and susceptibility-by-movements artifacts by Taylor expanding the measured field to approximate the effect of movement on the field map9. The systematic joint correction is carried out with FSL’s eddy_cuda function.
Self-supervised learning based denoising
To increase the low SNR inherent in high resolution infant dMRI, the eddy current and motion corrected DWI volumes undergo Patch2Self self-supervised learning-based denoising in which a regressor model is trained on patches in (n-1) DWI volumes to predict voxel-wise intensities in the nth held-out volume10. The Patch2Self takes advantage of the fact that the noise is a random fluctuation and does not make additional strong assumptions about the structure of the signal. To ensure enough computer memory for the memory-intense denoising, denoising is done on partial brain (10 slices) at a time.
High fidelity Diffusion tensor and kurtosis fitting
Affine registration of b=1000s/mm2 DWI volumes is carried out before diffusion tensor fitting in DTIStudio11 to resolve residual misalignment in the data and smooth out the data. Diffusion kurtosis fitting is directly done on partial brain denoised data in Dipy12.
Results
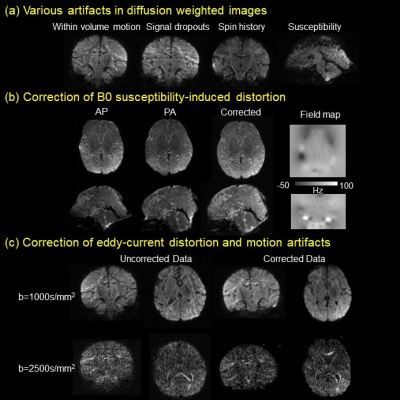
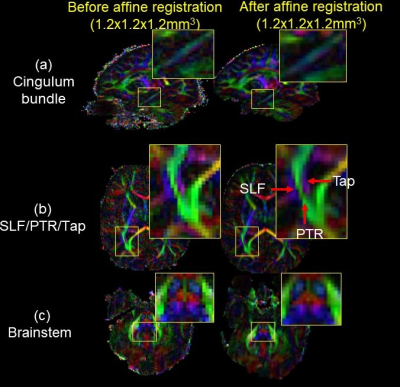
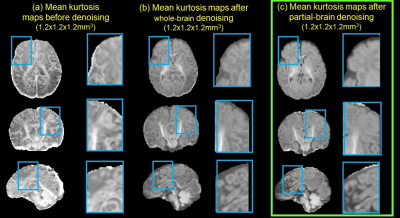
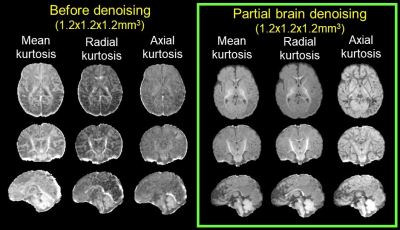
Fig. 2a demonstrates various artifacts in DWI common in high-resolution infant brain dMRI. Fig. 2b shows correction of B0 susceptibility-induced distortion with both phase encoding directions as inputs and the corrected b0, along with the estimated field map. Fig. 2c shows correction of eddy-current and other motion artifacts including within-volume motion and susceptibility-by-movement artifacts caused by head rotations. Corrected data show improved slice-to-volume alignment, reduced signal drop out, and significantly reduced image distortion. Fig. 3 demonstrates how 1.2mm isotropic resolution can help differentiate small tracts. Red arrows (Fig. 3b) point to three well-resolved tracts that are very close to each other, the superior longitudinal fasciculus (SLF), posterior thalamic radiation (PTR), and the tapetum (Tap). Affine registration also improves tract contrasts for the cingulum bundle, SLF/PTR/Tap, and brainstem in the orientation-encoded colormap as show in enlarged images (Fig. 3). Denoising significantly improves the fidelity and SNR of estimated mean kurtosis (MK) maps (Fig. 4). Fitted MK maps before denoising, with whole brain denoising which suffers from computer memory issue, and with partial brain denoising free from computer memory issues show gradual improvement in fidelity and SNR. Partial brain denoising (green box) demonstrates clearly improved contrasts among cerebral cortex, white matter and subcortical deep gray matter with improved SNR. In contrast, MK maps fitted before denoising and with whole brain denoising cannot clearly demonstrate high fidelity contrasts and are corrupted by noise. In Fig. 5, additional high fidelity radial kurtosis (RK) and axial kurtosis (AK) maps show high RK in the deep WM with radial restricted diffusion and low AK in deep WM where water diffusion freely along the axons (axial direction). MK, RK, AK contrasts before denoising demonstrate low SNR and low fidelity by contrast.Conclusion
We have developed a robust image analysis pipeline for high fidelity kurtosis and tensor fitting of 1.2mm isotropic infant brain diffusion MRI. Estimated high resolution diffusion tensor and kurtosis maps show high fidelity and high SNR relatively free from the severe motion artifacts and inherent low SNR in infant brain dMRI. Our robust pipeline allows us to efficiently analyze in-vivo dMRI data at 1.2mm resolution despite the considerable technical challenges specific to infant imaging.Acknowledgements
This study is funded by NIH R01MH092535, R01MH125333, R01EB031284, R21MH123930 and P50HD105354.References
1. Ouyang, M., Dubois, J., Yu, Q., Mukherjee, P., & Huang, H. Delineation of early brain development from fetuses to infants with diffusion MRI and beyond. 2019; Neuroimage, 185, 836-850.
2. Bastiani, M., Andersson, J. L., Cordero-Grande, L., Murgasova, M., Hutter, J., Price, A. N., ... & Sotiropoulos, S. N. Automated processing pipeline for neonatal diffusion MRI in the developing Human Connectome Project. 2019; Neuroimage, 185, 750-763.
3. Makropoulos, A., Robinson, E. C., Schuh, A., Wright, R., Fitzgibbon, S., Bozek, J., ... & Rueckert, D. The developing human connectome project: A minimal processing pipeline for neonatal cortical surface reconstruction. 2018; Neuroimage, 173, 88-112.
4. Harms, M. P., Somerville, L. H., Ances, B. M., Andersson, J., Barch, D. M., Bastiani, M., ... & Yacoub, E. Extending the Human Connectome Project across ages: Imaging protocols for the Lifespan Development and Aging projects. 2018; Neuroimage, 183, 972-984.
5. Andersson, J. L. & Sotiropoulos, S. N. Non-parametric representation and prediction of single- and multi-shell diffusion-weighted MRI data using Gaussian processes. 2015; Neuroimage 122, 166-176
6. Andersson, J. L. & Sotiropoulos, S. N. An integrated approach to correction for off-resonance effects and subject movement in diffusion MR imaging.2016; Neuroimage 125, 1063-1078
7. Andersson, J. L., Graham, M. S., Zsoldos, E. & Sotiropoulos, S. N. Incorporating outlier detection and replacement into a non-parametric framework for movement and distortion correction of diffusion MR images.2016; Neuroimage 141, 556-572
8. Andersson, J. L., Graham, M. S., Drobnjak, I., Zhang, H., Filippini, N., & Bastiani, M. Towards a comprehensive framework for movement and distortion correction of diffusion MR images: Within volume movement. 2017; Neuroimage, 152, 450-466.
9. Andersson, J. L., Graham, M. S., Drobnjak, I., Zhang, H., & Campbell, J. Susceptibility-induced distortion that varies due to motion: Correction in diffusion MR without acquiring additional data.2018; Neuroimage, 171, 277-295.
10. Fadnavis, S., Batson, J., & Garyfallidis, E. Patch2Self: Denoising Diffusion MRI with Self-Supervised Learning. 2020; Advances in Neural Information Processing Systems, 33, 16293-16303.
11. Jiang, H., Van Zijl, P. C., Kim, J., Pearlson, G. D., & Mori, S. DtiStudio: resource program for diffusion tensor computation and fiber bundle tracking. 2006; Computer methods and programs in biomedicine, 81(2), 106-116.
12. Garyfallidis, E., Brett, M., Amirbekian, B., Rokem, A., Van Der Walt, S., Descoteaux, M., ... & Dipy Contributors. Dipy, a library for the analysis of diffusion MRI data. 2014; Frontiers in neuroinformatics, 8
Figures