3115
Self-Supervised Image Reconstruction of 7T MP2RAGE for Multiple Sclerosis: 0.5mm Isotropic Resolution in 10 Minutes1Advanced Clinical Imaging Technology, Siemens Healthineers International AG, Lausanne, Switzerland, 2Signal Processing Laboratory 5 (LTS5), Ecole Polytechnique Fédérale de Lausanne (EPFL), Lausanne, Switzerland, 3Department of Radiology, Lausanne University Hospital and University of Lausanne, Lausanne, Switzerland, 4Department of Neurology, Icahn School of Medicine at Mount Sinai, New York, NY, United States, 5Centre d’Imagerie Biomédicale (CIBM), EPFL, Lausanne, Switzerland, 6Human Neuroscience Platform, Fondation Campus Biotech Geneva, Geneva, Switzerland, 7National Institute of Neurological Disorders and Stroke, National Institutes of Health, Bethesda, MD, United States, 8Siemens Healthineers International AG, Erlangen, Germany
Synopsis
Keywords: Image Reconstruction, Machine Learning/Artificial Intelligence, Ultra-High Field MRI
High-resolution 3D MR imaging is necessary for the detailed assessment of focal pathologies, such as cortical lesions. However, high-resolution demands tradeoffs with acceleration and SNR, which is difficult to address with standard machine learning reconstructions due to the infeasibility of collecting large datasets of fully sampled data. Using a dataset of high-resolution (0.5mm isotropic), 3D, 7T MP2RAGE scans of multiple sclerosis patients, we show that a self-supervised reconstruction from one scan, requiring no fully sampled data, has higher apparent SNR than a median of three scans, currently used for assessment, with comparable tissue contrast and lesion conspicuity.Introduction
In MRI, acquisitions need to balance acceleration, SNR, and spatial resolution, with higher resolution typically traded for lower intrinsic SNR. While deep learning (DL) methods in MR image reconstruction have resulted in unprecedented levels of image quality from highly undersampled data1, such methods tend to require large amounts of ground truth, fully sampled data, the collection of which is impractical or even infeasible due to long acquisition times for 3D and high-resolution acquisitions.In this abstract, we show, in an important clinical application, that this tradeoff can potentially be circumvented using only prospectively accelerated scans; we show that a state of the art, self-supervised reconstruction method, which has been empirically and theoretically shown to accurately denoise and reconstruct images2,3, can increase the image SNR from a fixed, high-resolution acquisition solely by training on undersampled data with no access to ground truth, fully sampled data.
We validate this with a dataset of high-resolution, 3D, 7T MP2RAGE4 acquisitions using a protocol where multiple sclerosis5 (MS) patients were scanned three times in a single session and a median image was computed to facilitate assessment of cortical lesions6,7. We compare the image quality and lesion conspicuity of self-supervised reconstructions and the median images, quantitatively and qualitatively.
Methods
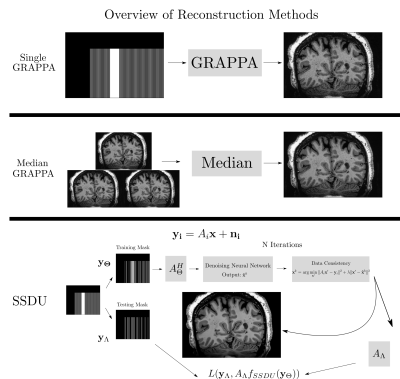
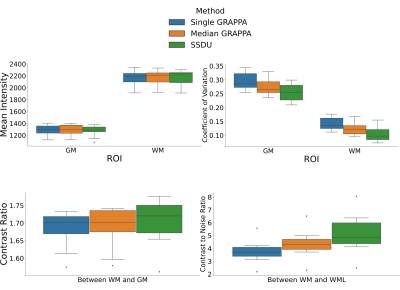
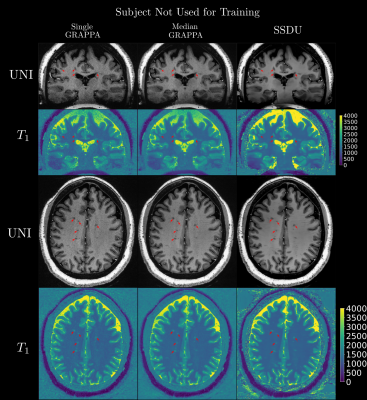
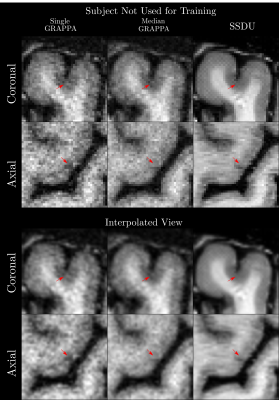
Seven subjects with MS and two healthy volunteers (HV) were scanned three times in a single session using a 3D MP2RAGE research application sequence (TR/TI1/TI2/TE = 6000/800/2700/5 ms, voxel size = 0.5×0.5×0.5 mm3, matrix size=224x336x448, acquisition time=10:30 min) with 6/8 Partial Fourier acceleration in both phase encoding directions and parallel imaging acceleration of 3 along one phase encoding direction. We compare three image reconstruction methods: a GRAPPA8 reconstruction of the first scan, the median of GRAPPA reconstructions from all three scans (co-registered to the first scan), and a recently proposed, self-supervised reconstruction of the first scan (see Figure 1), called Self-Supervision via Data Undersampling (SSDU)9. SSDU consists of an un-rolled network that iteratively applies a denoising step, through a neural network, and a data consistency step through conjugate gradient optimization. The un-rolled network is trained solely on the undersampled data from the first scan of 5 MS patients and the two HV (2D, coronal slices generated from inverse Fourier transforming the data along the readout direction). As the approach is self-supervised, the trained models are, a priori, specific to the subjects used in training, and we perform inference on the same data for comparison; however, as a robustness check, we show qualitative comparison on subjects not seen in training, which exhibited higher noise/motion than those in the training set. A separate model is trained for both MP2RAGE inversion contrasts, with the two images being combined after inference to form the uniform (UNI) image without background noise10.For quantitative analysis, an automatic tissue segmentation from an in-house method based on nnUNet11 was used to segment gray matter (GM), white matter (WM) and WM lesions (WML). The segmentation was performed on the median GRAPPA image and propagated to the other methods. We compared the mean intensity and the coefficient of variation (COV) of the intensity in WM and GM, the contrast ratio (CR) between WM and GM, and the contrast to noise ratio (CNR) between WM and WML in the UNI images over all nine subjects. The CNR is computed using the standard deviation in WM as the noise estimate. For qualitative comparison, we show example slices of MP2RAGE T1-weighted UNI images and T1 maps as well as closeups of CL which were manually segmented.
Results
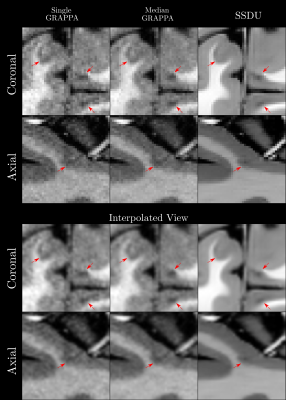
Quantitatively (Figure 2), we found that while all methods had comparable intensities in the ROIs, SSDU generally had the lowest COV, indicating superior denoising. This is supported by comparable CR between WM and GM amongst all methods and higher CNR between WM and WML for SSDU, indicating that tissue contrast is preserved, despite denoising.Qualitatively (Figure 3), using a subject not used for training, we can see that SSDU provides a smoother reconstruction than the other methods while still preserving edges and contrast between different tissues; furthermore, WML can still be clearly delineated. Additionally, Figures 4-5 show that small CL can be seen on SSDU reconstructions, albeit potentially with some oversmoothing.
Discussion and Conclusion
The results show that SSDU can reconstruct images with higher SNR and comparable tissue contrast and lesion conspicuity from a single scan, compared to the median of three scans.However, as SSDU is trained/reconstructs the 3D volumes slice wise (coronal), related stripe artifacts appear in the axial and sagittal views; more work is required to avoid these. While we can regard the trained models as specific to the subjects in the training set, the generalizability of the trained model to subjects not used for training should be further investigated in more subjects as this could save future computation/deployment time. Finally, while the results are promising, as this is a preliminary study with few participants, more extensive validation12 is necessary, including rigorous comparisons of automatic/manual segmentations using images from the different methods.
In conclusion, the results show the potential for self-supervised reconstructions to facilitate high resolution and high SNR in a reasonable acquisition time, without having access to fully sampled data.
Acknowledgements
F.L.R. is supported by the Swiss National Science Foundation (SNF) Postdoc Mobility Fellowship (P500PB_206833)
TY,GFP,PL,TK,TH are employed by Siemens Healthineers International AG.
References
1. Knoll, Florian, Kerstin Hammernik, Chi Zhang, Steen Moeller, Thomas Pock, Daniel K. Sodickson, and Mehmet Akcakaya. "Deep-learning methods for parallel magnetic resonance imaging reconstruction: A survey of the current approaches, trends, and issues." IEEE signal processing magazine 37, no. 1 (2020): 128-140.
2. Batson, Joshua, and Loic Royer. "Noise2self: Blind denoising by self-supervision." In International Conference on Machine Learning, pp. 524-533. PMLR, 2019.9.
3. Yu, Thomas, Tom Hilbert, Gian Franco Piredda, Arun Joseph, Gabriele Bonanno, Salim Zenkhri, Patrick Omoumi et al. "Validation and Generalizability of Self-Supervised Image Reconstruction Methods for Undersampled MRI." arXiv preprint arXiv:2201.12535 (2022).
4. Marques, José P., Tobias Kober, Gunnar Krueger, Wietske van der Zwaag, Pierre-François Van de Moortele, and Rolf Gruetter. "MP2RAGE, a self bias-field corrected sequence for improved segmentation and T1-mapping at high field." Neuroimage 49, no. 2 (2010): 1271-1281.
5. Thompson, Alan J., Brenda L. Banwell, Frederik Barkhof, William M. Carroll, Timothy Coetzee, Giancarlo Comi, Jorge Correale et al. "Diagnosis of multiple sclerosis: 2017 revisions of the McDonald criteria." The Lancet Neurology 17, no. 2 (2018): 162-173.
6. Beck, Erin S., Pascal Sati, Varun Sethi, Tobias Kober, Blake Dewey, Pavan Bhargava, Govind Nair, Irene C. Cortese, and Daniel S. Reich. "Improved visualization of cortical lesions in multiple sclerosis using 7T MP2RAGE." American Journal of Neuroradiology 39, no. 3 (2018): 459-466.
7. Beck, Erin S., Josefina Maranzano, Nicholas J. Luciano, Prasanna Parvathaneni, Stefano Filippini, Mark Morrison, Daniel J. Suto et al. "Cortical lesion hotspots and association of subpial lesions with disability in multiple sclerosis." Multiple Sclerosis Journal (2022): 13524585211069167.
8. Griswold, Mark A., Peter M. Jakob, Robin M. Heidemann, Mathias Nittka, Vladimir Jellus, Jianmin Wang, Berthold Kiefer, and Axel Haase. "Generalized autocalibrating partially parallel acquisitions (GRAPPA)." Magnetic Resonance in Medicine: An Official Journal of the International Society for Magnetic Resonance in Medicine 47, no. 6 (2002): 1202-1210.
9. Yaman, Burhaneddin, Seyed Amir Hossein Hosseini, Steen Moeller, Jutta Ellermann, Kâmil Uğurbil, and Mehmet Akçakaya. "Self‐supervised learning of physics‐guided reconstruction neural networks without fully sampled reference data." Magnetic resonance in medicine 84, no. 6 (2020): 3172-3191.
10. O'Brien KR, Kober T, Hagmann P, Maeder P, Marques J, Lazeyras F, Krueger G, Roche A. Robust T1-weighted structural brain imaging and morphometry at 7T using MP2RAGE. PLoS One. 2014 Jun 16;9(6):e99676. doi: 10.1371/journal.pone.0099676. PMID: 24932514; PMCID: PMC4059664.
11. Isensee, Fabian, Paul F. Jaeger, Simon AA Kohl, Jens Petersen, and Klaus H. Maier-Hein. "nnU-Net: a self-configuring method for deep learning-based biomedical image segmentation." Nature methods 18, no. 2 (2021): 203-211.
12. Yamamoto, T., C. Lacheret, H. Fukutomi, R. A. Kamraoui, L. Denat, B. Zhang, V. Prevost et al. "Validation of a Denoising Method Using Deep Learning–Based Reconstruction to Quantify Multiple Sclerosis Lesion Load on Fast FLAIR Imaging." American Journal of Neuroradiology 43, no. 8 (2022): 1099-1106.
13. Pruessmann, Klaas P., Markus Weiger, Markus B. Scheidegger, and Peter Boesiger. "SENSE: sensitivity encoding for fast MRI." Magnetic Resonance in Medicine: An Official Journal of the International Society for Magnetic Resonance in Medicine 42, no. 5 (1999): 952-962.
14. Vaswani, Ashish, Noam Shazeer, Niki Parmar, Jakob Uszkoreit, Llion Jones, Aidan N. Gomez, Łukasz Kaiser, and Illia Polosukhin. "Attention is all you need." Advances in neural information processing systems 30 (2017).
Figures