3112
Accelerating 7T susceptibility-weighted imaging with complex-valued convolutional neural network1Department of Radiology, Chinese PLA General Hospital, Beijing, China, 2MR Collaboration, Siemens Healthineers Ltd., Beijing, China, 3Key Laboratory of Magnetic Resonance in Biological Systems, State Key Laboratory of Magnetic Resonance and Atomic and Molecular Physics, National Center for Magnetic Resonance in Wuhan, Wuhan Institute of Physics and Mathematics, Innovation Academy for Precision Measurement Science and Technology, Chinese Academy of Sciences‒Wuhan National Laboratory for Optoelectronics, Wuhan, China
Synopsis
Keywords: Image Reconstruction, Machine Learning/Artificial Intelligence
Ultra-high field 7T susceptibility-weighted imaging (SWI) has shown great potential in visualizing and evaluating a broad range of pathology, but suffers from long acquisition times. In this study, a complex-valued convolutional neural network (ComplexNet) model was proposed to reconstruct highly accelerated 7T SWI data. The average reconstruction time of ComplexNet was 0.56 seconds per slice (45.16 seconds per participant). Meanwhile, ComplexNet can provide high-quality 7T SWI for visualizing subtle pathology, including cerebral microbleeds, asymmetric deep medullary veins, and swallow tail sign.Introduction
At ultra-high field 7T MRI, SWI can provide superior depiction of venous microvasculature, cerebral microbleeds, or deep gray matter structures (e.g., substantia nigra, subthalamic nucleus, and thalamus).1 However, 7T SWI suffers from long scan times (usually 7-10 minutes with parallel imaging) due to sub-millimeter resolution acquisitions and long echo time.2 The long acquisition time can easily lead to motion artifacts even in compliant subjects, and impose great burdens on patients with brain diseases. In this study, a complex-valued convolutional neural network (ComplexNet) was developed to fast and accurately reconstruct highly accelerated 7T SWI data, and its performance in terms of reconstruction quality and pathology visualization was investigated.Methods
To faithfully reconstruct both MR magnitude and phase images, a deep learning model based on ComplexNet was proposed in this work.3 ComplexNet consists of a deep cascade of convolutional neural network (CNN) modules and data consistency layers. We adopt the complex residual network as the CNN modules to leverage the inherently complex-valued nature of SWI data and learn richer representations. Furthermore, each CNN module is followed by a data consistency layer to ensure data consistency in k-space domain. The mean-squared error between the ComplexNet reconstructed results and the fully sampled images was chosen as the loss function.The SWI data were acquired at a whole-body 7T scanner (MAGNETOM Terra, Siemens Healthcare, Erlangen, Germany) equipped with a 32-channel head coil (Nova Medical, Wilmington, MA, USA) using a flow-compensated 3D SWI sequence. Imaging parameters were as follows: repetition time = 21.0 ms, echo time = 14.0 ms, field of view = 220 × 179 mm2, slice thickness = 1.5 mm, voxel size = 0.25 × 0.25 × 1.5 mm3, flip angle = 15°, bandwidth = 210 Hz/Px, number of slices = 80, 3 × undersampling in the first phase-encoding direction, and a total acquisition time of approximately 7 minutes 29 seconds.
The SWI data were acquired from 130 participants, including patients with tumor, cerebrovascular disease, Parkinson’s disease, multiple sclerosis, and other conditions. The dataset was split into 2D axial slices of 91 participants for training, 39 participants for testing. The undersampled data were generated by retrospectively undersampling the reference k-space data with acceleration rates (R) of 6 and 8. The reconstruction results of ComplexNet were evaluated using two quantitative metrics: peak signal-to-noise ratio (PSNR) and structural similarity (SSIM).
Results
Mean PSNR values were 39.48 ± 2.17 and 38.79 ± 1.92 for ComplexNet at R = 6 and R = 8, respectively. Mean SSIM values were 0.9563 ± 0.0233 and 0.9517 ± 0.0235 for ComplexNet at R = 6 and R = 8, respectively. Average reconstruction time was 0.56 seconds per slice (45.16 seconds per participant) for ComplexNet in TensorFlow on the RTX 3090 GPU.Figure 1 shows representative SWI images obtained using fully sampled and ComplexNet approaches at R = 6 and R =8 in a 63-year-old woman with Parkinson’s disease. It can be seen that the swallow tail sign is not clearly shown in the fully sampled images (Figure 1a) due to the degeneration of nigrosome 1 in PD. This potential imaging biomarker can also be consistently identified in the SWI images obtained using ComplexNet at both R = 6 (Figure 1b) and R = 8 (Figure 1c). In Figure 2, ComplexNet shows a similar diagnostic image quality as the fully sampled approach for visualization of subtle pathology, including asymmetric deep medullary veins, cerebral venous malformation, and multiple cerebral microbleeds.
Discussion and conclusion
SWI particularly benefits from increased SNR as well as the enhanced sensitivity of susceptibility effects at higher field strengths.4 Therefore, high-resolution SWI at 7T is a powerful tool to improve lesion characterization and treatment planning. In this study, we developed a ComplexNet model for fast and accurate reconstruction of highly accelerated 7T SWI data. Our results show that ComplexNet can offer comparable performance to the fully sampled SWI for visualizing a wide range of pathology. In a prospective way, the scan time of 7T SWI could be reduced by approximately 83.3% for R = 6, and 87.5% for R = 8 compared to the fully sampled acquisition, which is of greatest interest regarding clinical practice.Acknowledgements
This work is supported by the National Natural Science Foundation of China (81825012, 81730048 and 82151309).References
[1] Rutland J W, Delman B N, Gill C M, et al. Emerging use of ultra-high-field 7T MRI in the study of intracranial vascularity: state of the field and future directions. AJNR Am J Neuroradiol. 2020;41:2–9.
[2] Jorge J, Gretsch F, Najdenovska E, et al. Improved susceptibility-weighted imaging for high contrast and resolution thalamic nuclei mapping at 7T. Magn Reson Med. 2020;84:1218–1234.
[3] Duan C, Xiong Y, Cheng K, et al. Accelerating susceptibility-weighted imaging with deep learning by complex-valued convolutional neural network (ComplexNet): validation in clinical brain imaging. Eur Radiol. 2022;32:5679–5687.
[4] Deistung A, Rauscher A, Sedlacik J, et al. Susceptibility weighted imaging at ultra high magnetic field strengths: Theoretical considerations and experimental results. Magn Reson Med. 2008;60:1155-1168.
Figures
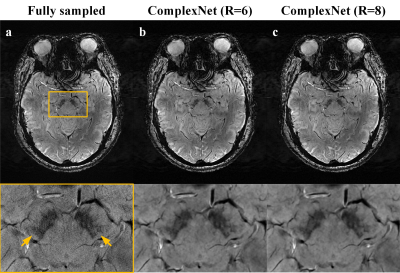
Figure 1. Representative SWI images comparing the fully sampled and ComplexNet approaches at R = 6 and R = 8 for visualization of swallow tail sign.
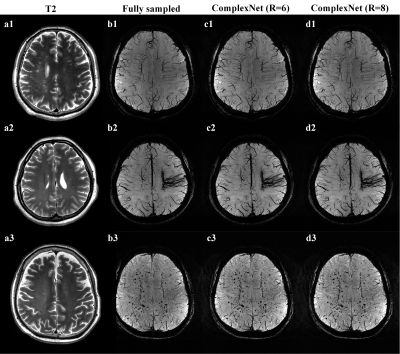
Figure 2. Minimum intensity projection of SWI data obtained using fully sampled and ComplexNet approaches, with T2-weighted MR images (a1–a3) shown as reference. a1–d1 Images in a 65-year-old man with bilateral severe carotid artery stenosis. a2–d2 Images in a 26-year-old woman with cerebral venous malformation. a3–d3 images in a 54-year-old man with multiple cerebral microbleeds.