3104
Task-based evaluation of deep learning-based reconstruction for highly-accelerated 3D T1-weighted brain MRI scans1GE Research, Niskayuna, NY, United States, 2Mayo Clinic College of Medicine, Rochester, MN, United States, 3Walter Reed National Military Medical Center, Bethesda, MD, United States, 4Uniformed Services University of the Health Sciences, Bethesda, MD, United States, 5GE Research, Herzliya, Israel, 6GE HealthCare, Waukesha, WI, United States, 7GE HealthCare, Rochester, MN, United States, 8GE HealthCare, Menlo Park, CA, United States
Synopsis
Keywords: Image Reconstruction, Machine Learning/Artificial Intelligence, Brain, Neuro
3D MRI enables thin slices at the cost of long scan times, causing practical challenges. Recently, deep-learning (DL) techniques have successfully accelerated MR scans. However, it is challenging to characterize the image quality (IQ) performance of DL methods by conventional metrics because IQ depends on applications, i.e., how images are used. We evaluate the IQ performance of DL-Speed, our DL-based acceleration method, for 3D T1-weighted MPRAGE brain scans, in 1) post-reconstruction subcortical structure segmentation, and 2) a reader study. The results imply DL-Speed can accelerate scans with reduction factor R=10 while maintaining IQ comparable to standard parallel imaging with R=2.1.Introduction
Three-dimensional (3D) MRI has advantages over 2D MRI, including higher isotropic spatial resolution achievable with thinner slices, and higher signal-to-noise ratio (SNR). However, the benefits of 3D MRI come at the cost of longer scan times, which makes it susceptible to motion-related artifacts. To accelerate MRI scans, a variety of compressed-sensing and deep-learning (DL) techniques1 have been used for reconstructing images from highly-undersampled data. However, the maximum practical acceleration realized was about 6x2-5. Our version of DCI-Net (Densely Connected Iterative Network) reconstruction6,7, referred to as DL-Speed, is an unrolled iterative reconstruction method that can achieve up to 10x acceleration. Performance metrics for evaluating DL acceleration include scan time and image quality. However, it is challenging to assess image quality because it should reflect how reconstructed images are used. Conventional metrics, such as the mean-squared error and the structural similarity index measure (SSIM), would not properly measure task-dependent performance. In this study, we evaluate our DL-based acceleration method, DL-Speed, in 10x accelerated 3D T1-weighted MPRAGE brain scan, using two approaches. First, the impact of DL-Speed on brain segmentation is studied. Segmentation of subcortical brain structures is crucial to the study of various neurological disorders8. We test if DL-Speed from highly-undersampled MPRAGE data would yield subcortical structure segmentation results comparable to standard parallel imaging, using a deformable atlas-based segmentation method9. Second, diagnostic image quality for DL-Speed is evaluated by radiologists. While retrospectively-undersampled images were assessed in our previous study7,10, we evaluate prospectively-undersampled DL-Speed images in this study. In summary, we test if 3D MPRAGE can be 10x accelerated by DL-Speed while comparable to clinical parallel imaging for subcortical structure segmentation and neuroradiologist image assessment.Methods
3D sagittal MPRAGE data with 1-mm isotropic resolution were acquired on a high-performance 3.0 T MAGNUS head-gradient system (200 mT/m and 500 T/m/s gradients)11 using a 32-channel receive coil (NOVA Medical, Wilmington, MA, USA). Eight healthy subjects were recruited under an IRB-approved protocol and provided written informed consent. For each subject, prospectively-undersampled MPRAGE data with an acceleration factor of R=10.2±0.2 (mean ± standard deviation) with a variable density Poisson disc (VD-PD)12 sampling pattern were acquired (scan time: 67±0.4 s). The undersampled data were reconstructed using our DCI-Net6,7,10, that is, DL-Speed (Figure 1). As a baseline, standard parallel imaging data with R=2.1±0.1 with a regular sampling pattern were acquired (scan time: 320±16 s) and reconstructed using Autocalibrating Reconstruction for Cartesian imaging (ARC)13 (see Figure 2 for representative images). In addition, fully-sampled data were generated from the parallel imaging data using ARC and then retrospectively undersampled using the same VD-PD sampling pattern. The retrospectively-undersampled data were also reconstructed using DCI-Net.The DL-Speed images and baseline ARC images were segmented for 6 subcortical structures (left and right, separately) including hippocampus, thalamus, caudate nucleus, putamen, pallidum and amygdala, using the deformable atlas-based segmentation method9 for all 8 subjects. The volumes of the segmented structures were compared for DL-Speed and ARC. In addition, Dice coefficients between segmented structure volumes for DL-Speed and ARC were calculated.
For 3 subjects, prospectively-undersampled DL-Speed images (R=10.4, scan time: 67 s) and ARC images (R=2.1, scan time: 306 s) were assessed by 3 board-certified neuroradiologists blinded to sequence and reconstruction. Image scoring was based on consensus reading, on a 5-point Likert scale (1: poor, 5: excellent), for 8 categories including SNR, artifacts, gray/white matter contrast, resolution/sharpness, deep gray, cerebellar vermis, anterior commissure, and overall quality.
Results
Figures 3 and 4 show the relative volume difference and the Dice coefficient between ARC (R=2.1±0.1) and retrospectively-undersampled DL-Speed (R=10.2±0.2), respectively, averaged over all 8 subjects, for each subcortical structure.Figure 5 shows the image scores averaged over the 3 subjects for ARC (R=2.1, scan time: 306 s) and prospectively-undersampled DL-Speed (R=10.4, scan time: 67 s) from the reader study.
Discussion
DL-Speed (R=10.2) yielded subcortical structure segmentation results comparable to ARC (R=2.1), that is, volume difference ≤1.3% except for pallidum (Figure 3) and reasonably high overlap (dice coefficients ≥ 0.94) (Figure 4). The relative volume difference was higher (3.5%) for pallidum (Figure 3) probably because the pallidum is small and the deep grey region has lower SNR, which requires further investigation and optimization of DL-Speed. The reader study implies prospectively-undersampled DL-Speed (R=10.4, scan time: 67 s) provides diagnostically acceptable image quality (scores ≥ 4) although the scores are slightly lower than those for ARC (R=2.1, scan time: 306 s) (Figure 5).Although the results are promising, the number of subjects (8 for subcortical structure segmentation, and 3 for reader study) is small. We plan to perform larger-scale studies. In this study, for segmentation, we used the atlas-based method9 that we implemented, and we focused on a comparison of segmentation results from ARC and DL-Speed rather than on the segmentation performance. It would be worthwhile to test other brain segmentation methods8. Although we fixed the DL-Speed model in this study, there is room for improvement in the image quality performance by optimizing the network, training data, and sampling patterns.
Conclusion
DL-Speed enables a 10-fold acceleration of 3D T1-weighted MPRAGE brain scans, which amounts to about a 4.8-fold acceleration with respect to the standard parallel imaging (R=2.1), while maintaining comparable subcortical structure segmentation and diagnostic image quality.Acknowledgements
This work was supported in part by CDMRP under Grant W81XWH-16-2-0054 and by NIH under Grant U01EB024450.References
1. Sandino CM, Cheng JY, Chen F, et al. Compressed sensing: from research to clinical practice with deep neural networks. IEEE Signal Process Mag. 2020;37(1):111-127.
2. Li Z, Tian Q, Ngamsombat C, et al. High-fidelity fast volumetric brain MRI using synergistic wave-controlled aliasing in parallel imaging and a hybrid denoising generative adversarial network (HDnGAN). Med Phys. 2022;49(2):1000-1014.
3. Cristobal-Huerta A, Poot DHJ, Vogel MW, et al. Compressed sensing 3D-GRASE for faster high-resolution MRI. Magn Reson Med. 2019;82(3):984-999.
4. Delattre BMA, Boudabbous S, Hansen C, et al. Compressed sensing MRI of different organs: ready for clinical daily practice? Eur Radiol. 2020;30(1):308-319.
5. Duan Y, Zhang J, Zhuo Z, et al. Accelerating brain 3D T1-weighted turbo field echo MRI using compressed sensing-sensitivity encoding (CS-SENSE). Eur J Radiol. 2020;131:109255.
6. Malkiel I, Ahn S, Slavens Z, et al. Densely connected iterative network for sparse MRI reconstruction. ISMRM 2018, p. 3363.
7. Ahn S, Wollner U, McKinnon G, et al. Deep learning-based reconstruction of highly accelerated 3D MRI. arXiv: 2203.04674, 2022.
8. Dolz J, Desrosiers C, Ayed IB. 3D fully convolutional networks for subcortical segmentation in MRI: A large-scale study. NeuroImage. 2018;170:456-470.
9. Liu X, Montillo A, Tan ET, et al. Deformable atlas for multi-structure segmentation. Medical Image Computing and Computer-Assisted Intervention - MICCAI 2013, Lecture Notes in Computer Science. 2013;8149:743-750.
10. Ahn S, Wollner U, McKinnon G, et al. Deep learning-based reconstruction of highly-accelerated 3D MRI MPRAGE images. ISMRM 2021.
11. Foo TKF, Tan ET, Vermilyea ME, et al. Highly efficient head‐only magnetic field insert gradient coil for achieving simultaneous high gradient amplitude and slew rate at 3.0T (MAGNUS) for brain microstructure imaging. Magn Reson Med. 2020;83(6):2356-2369.
12. Lugauer F, Wetzl J, Forman C, et al. Single-breath-hold abdominal T1 mapping using 3D Cartesian Look-Locker with spatiotemporal sparsity constraints. Magn Reason Mater Phy. 2018;31:399-414.
13. Brau AC, Beatty PJ, Skare S, et al. Comparison of reconstruction accuracy and efficiency among autocalibrating data-driven parallel imaging methods. Magn Reson Med. 2008;59(2):382-395.
14. Ahn S, Menini A, McKinnon G, et al. Contrast-weighted SSIM loss function for deep learning-based undersampled MRI reconstruction. ISMRM 2020.
Figures
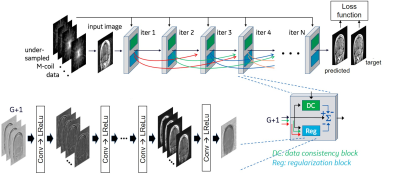
Figure 1. Architecture of DCI-Net6,7, referred to as DL-Speed. We used 28 iterations, 9 layers in each regularization block, 96 convolution kernels per layer, and up to 20 skip connections denoted by curved arrows. The network was trained using T1-weighted 3D brain scan data (133 datasets for training and 15 for validation) with retrospective undersampling (reduction factor, R=10) with variable-density Poisson-disc (VD-PD) sampling patterns. A contrast-weighted SSIM14 extended to complex-valued images was used for the loss function.
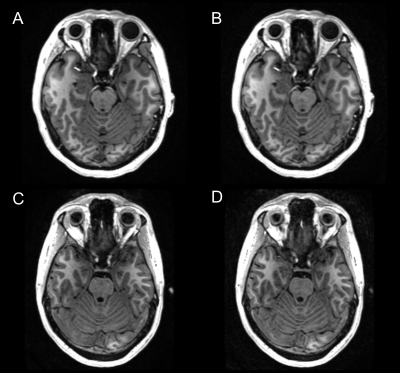
Figure 2. Representative ARC (R=2.1, scan time: 306 s) and prospectively-undersampled DL-Speed (R=10.4, scan time: 67 s) images for 3D T1-weighted MPRAGE with 1-mm isotropic resolution: (A) ARC and (B) DL-Speed images for subject 1, and (C) ARC and (D) DL-Speed images for subject 2. Note that ARC and prospectively-undersampled DL-Speed images are from different scans.
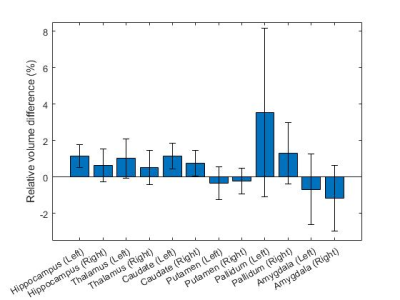
Figure 3. Relative volume difference between ARC (R=2.1±0.1) and retrospectively-undersampled DL-Speed (R=10.2±0.2) averaged over 8 subjects for each subcortical structure. The error bars denote 95% confidence intervals.
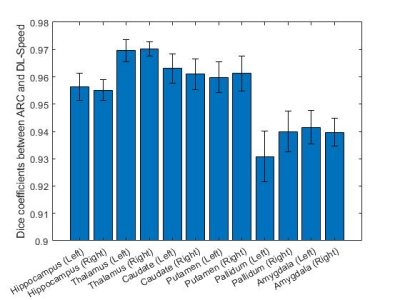
Figure 4. Dice coefficient between ARC (R=2.1±0.1) and retrospectively-undersampled DL-Speed (R=10.2±0.2) averaged over 8 subjects for each subcortical structure. The error bars denote 95% confidence intervals.
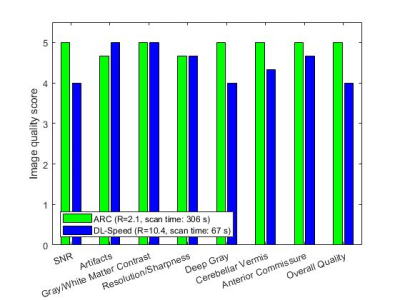
Figure 5. Image quality scores for ARC (R=2.1, scan time: 306 s) and prospectively-undersampled DL-Speed (R=10.4, scan time: 67 s) averaged over 3 subjects from reader study.