3100
Deep Magnetic Resonance Fingerprinting Based on local and global vision transformer
Peng Li1, Xiaodi Li1, Xin Lu2, and Yue Hu1
1Harbin Institute of Technology, Harbin, China, 2De Montfort University, Leicester, United Kingdom
1Harbin Institute of Technology, Harbin, China, 2De Montfort University, Leicester, United Kingdom
Synopsis
Keywords: Image Reconstruction, MR Fingerprinting
Magnetic resonance fingerprinting (MRF) can achieve simultaneous imaging of multiple tissue parameters. However, the size of the tissue fingerprint dictionary used in MRF grows exponentially as the number of tissue parameters increases, which may result in prohibitively large dictionaries that require extensive computational resources. Existing CNN-based methods obtain parameter reconstruction patch-wisely, using only local information and resulting in limited reconstruction speed. In this paper, we propose a novel end-to-end local and global vision transformer (LG-VIT) for MRF parameter reconstruction. The proposed method enables significantly fast and accurate end-to-end parameter reconstruction while avoiding the high computational cost of high-dimensional data.Introduction
Magnetic resonance fingerprinting (MRF) is a quantitative MRI method proposed by Ma et al.[1], which enables simultaneous imaging of multiple tissue parameters, including spin-lattice relaxation time ($$${\rm T}_1$$$), spin-spin relaxation time ($$${\rm T}_2$$$), etc. MRF uses a variable schedule of radiofrequency excitations and delays to induce unique signal evolutions from different tissues, so-called tissue fingerprints. The multiple quantization parameters are then obtained by mapping tissue fingerprints to a precomputed dictionary containing theoretical fingerprints of a set of tissues. To reduce errors in reconstructed parameter maps, the precomputed dictionary is usually computed with as little granularity as possible over the entire range of possible tissue values. However, the size of the tissue fingerprint dictionary used in MRF grows exponentially as the number of tissue parameters included is increased, which may quickly result in prohibitively large dictionaries that require extensive computational resources, e.g. memory, storage, computation time, etc.Recently, deep learning-based methods have been introduced as powerful alternatives to improve the speed and accuracy of parameter estimation in MRF. Cohen et al.[2] proposed a 4-layer fully connected neural network, termed as Drone, to perform signal-to-parameter mapping, replacing the memory-consuming dictionary and time-consuming dictionary matching. Soyak et al.[3] proposed a neural network consisting of a channel-wise attention module and a fully convolutional network (CONV-ICA) and adopted the strategy of overlapping patches for patch-level multi-parameter estimation to effectively reduce the error of parameter reconstruction. Fang et al.[4] proposed a two-step network (SCQ) for parameter map estimation, including a feature extraction module for reducing feature dimensionality and a spatially constrained quantification module for parameter map reconstruction. There are also many studies using different network structures for parametric map reconstruction. Most of the existing methods rely on CNN to capture features from MRF data and reconstruct parameters patch-wisely, resulting in a limited speed of reconstruction. However, CNN suffers from inherent locality limitations, making it difficult to capture long-distance correlations. In addition, it is difficult for 2D CNNs to capture temporal features, while deep 3D CNNs are extremely computationally expensive when dealing with high-dimensional MRF data.Methods
Fig.1 shows the overview of the proposed end-to-end local and global vision transformer network (LG-VIT). The high-dimensional data is first divided into patches and then fed into a local transformer encoder to parallelly extract lower-dimensional local features. Subsequently, a global transformer encoder is proposed to learn the global correlations among all patches. Finally, the multiple-parameter maps are reconstructed by using both local and global features. The local and global transformer encoder consists of $n$ transformer encoder blocks, which are shown in Fig.2. Each local transformer encoder block consists of a multi-head attention block and a multilayer perceptron (MLP) layer. To ensure the consistency of local and global position encodings, the position encodings are given by [5]. The proposed network utilizes both local and global encoders to fully capture the correlation features in both spatial and temporal dimensions of MRF data while avoiding the high computational cost of high-dimensional data. Our work was developed based on the Pytorch deep-learning framework and implemented on a Linux workstation with an Intel Xeon CPU and an Nvidia Quadro GV100 GPU.Results and discussion
The dataset used in this study consists of two parts: simulation data and in vivo data. The simulation data was generated by using the parametric map ($$${\rm T}_1$$$, $$${\rm T}_2$$$) taken from Brainweb[6]. The Bloch simulations were performed to generate the multi-frame MRF data by using the fast imaging with steady-state precession (FISP) pulse sequence. The in vivo data were acquired on a 3T Siemens Prisma scanner scanning 8 healthy volunteers across 10 slices each. Each volunteer was scanned using a 1000-length FISP sequence with a 32-channel head coil. The relevant imaging parameters include FOV = $$$220 \times 220$$$ $$${\rm mm}^2$$$, and slice thickness = 5 mm. All the undersampled data were first reconstructed using the SL-SP[7] method, followed by parameter estimation using each method. ~10% of the dataset is used for testing and the rest for training. We compare the performance of the proposed LG-VIT method with several state-of-the-art methods, including Drone[2], SCQ[4], and CONV-ICA[3]. We used the normalized mean square error (NMSE) to measure the quality of the reconstructed parametric maps. Fig.3 and Fig.4 show the parameter maps reconstructed by different methods from one set of simulated data and in vivo data in the test dataset, respectively. Table.1 shows quantitative comparison results of different methods on the test dataset. Experimental results show that our proposed method can reconstruct the parametric maps of MRF significantly faster and more accurately than other methods.Conclusion
We proposed a novel end-to-end local and global vision transformer (LG-VIT) for MRF parameter map reconstruction. Specifically, we proposed a local transformer encoder to extract lower-dimensional local features from high-dimensional patches in parallel. The local features were then fed into a global transformer encoder to learn the global correlation features across all local patch features. The proposed method enables significantly fast and accurate end-to-end parameter reconstruction while avoiding the high computational cost of high-dimensional data.Acknowledgements
This work is supported by China NSFC 61871159 and Natural Science Foundation of Heilongjiang YQ2021F005.References
- Dan Ma, Vikas Gulani, and et al., “Magnetic resonancefingerprinting,” Nature, vol. 495, no. 7440, pp. 187–192,2013.
- Ouri Cohen, Bo Zhu, and et al., “MR fingerprintingdeep reconstruction network (DRONE),” Magnetic resonancein medicine, vol. 80, no. 3, pp. 885–894, 2018.
- Refik Soyak, Ebru Navruz, and et al., “Channel AttentionNetworks for Robust MR Fingerprint Matching,”IEEE Transactions on Biomedical Engineering, vol. 69,no. 4, pp. 1398–1405, 2021.
- Zhenghan Fang, Yong Chen, and et al., “Deep learningfor fast and spatially constrained tissue quantification from highly accelerated data in magnetic resonancefingerprinting,” IEEE transactions on medical imaging,vol. 38, no. 10, pp. 2364–2374, 2019.
- Zelun Wang and Jyh-Charn Liu, “Translating math formulaimages to LaTeX sequences using deep neural networkswith sequence-level training,” International Journalon Document Analysis and Recognition (IJDAR),vol. 24, no. 1, pp. 63–75, 2021.
- D Louis Collins, Alex P Zijdenbos, and et al., “Designand construction of a realistic digital brain phantom,”IEEE transactions on medical imaging, vol. 17, no. 3,pp. 463–468, 1998.
- Yue Hu, Peng Li, and et al., “High-Quality MR FingerprintingReconstruction Using Structured Low-RankMatrix Completion and Subspace Projection,” IEEETransactions on Medical Imaging, vol. 41, no. 5, pp.1150–1164, 2021.
Figures
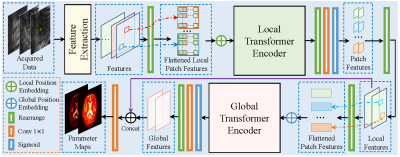
Fig.1 Overview of the proposed end-to-end local and global vision transformer network (LG-VIT).
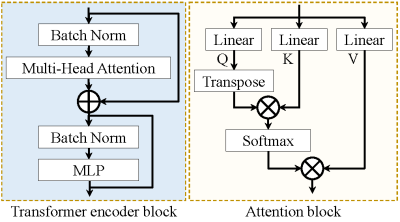
Fig.2 Illustration of the transformer encoder block and attention block.
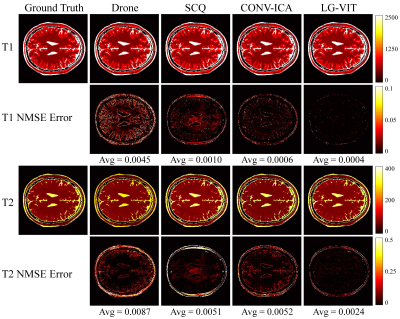
Fig.3 The parameter maps reconstructed by different methods from one set of simulated data in the test dataset. The first and second rows show the $$${\rm T}_1$$$ and $$${\rm T}_2$$$ parameter maps reconstructed by different methods, respectively. The third and fourth rows show the corresponding normalized mean squared error maps.
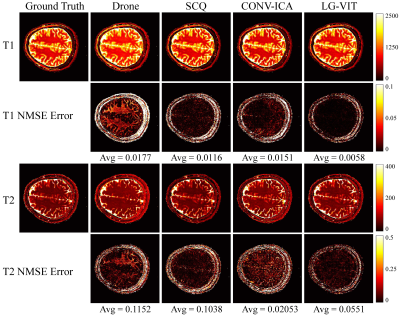
Fig.4 The parameter maps reconstructed by different methods from one set of in vivo data in the test dataset. The first and second rows show the $$${\rm T}_1$$$ and $$${\rm T}_2$$$ parameter maps reconstructed by different methods, respectively. The third and fourth rows show the corresponding normalized mean squared error maps.
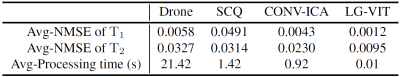
Table.1 Quantitative comparison of different methods on the test dataset.
DOI: https://doi.org/10.58530/2023/3100