3097
Effect of pathology on quantitative metrics of image reconstruction using a deep learning-based brain MRI reconstruction model
Shengjia Chen1, Patricia Johnson1, and Yvonne W. Lui1
1Department of Radiology, New York University Langone Health, New York, NY, United States
1Department of Radiology, New York University Langone Health, New York, NY, United States
Synopsis
Keywords: Image Reconstruction, Brain
We evaluate image quality in brain MR images with pathology, reconstructed by a deep learning-based image reconstruction algorithm. We have two main contributions: 1) a procedure for evaluating the image reconstruction quality of images, both globally and in patches with labelled pathology, and 2) report quantitative differences between two groups of reconstructed images (abnormal vs. normal). The pathology evaluation results find pathology regions have more losses and lower structural similarity when compared to normal patches and entire normal brains.Introduction
Recently, deep learning (DL) approaches have emerged as a complementary technique for accelerated MRI reconstruction quality with impressive results, particularly at high acceleration rates1-10. Currently, a major obstacle in the field is that DL reconstruction methods are often evaluated using global quantitative metrics such as structural similarity index (SSIM)11 or mean squared error (MSE), which tend to be well correlated with overall image quality but fail to measure the loss of fine detail or clinically important pathology12.In a previous study evaluating DL reconstruction in brain images, we observed what has been termed pseudonormalization of pathology at increasing acceleration rates and while it is felt that the rendering of abnormal findings likely suffers over that of more normal findings, quantitative measures for reconstruction quality over a large number of cases has not been done13. In this study, we quantify and compare reconstruction image quality in normal and abnormal (labelled pathology) cases on both a slice-based and regional basis.
Methods
DatasetsAll experiments were performed using the raw data from 5847 brain MRI belonging to the fastMRI brain dataset14-17. In addition, 1001 of these MRIs were labelled by subspecialist radiologist as part of the fastMRI+ project, and all focal pathology was labelled using a bounding box18. For model training and testing, labeled data were used as the test set (1,001 subjects) and unlabelled data were split between training (3,715 subjects) and validation sets (1,131 subjects). In the test set, we labeled slices having annotations in fastMRI+ as “abnormal” while the slices without annotations were labeled as normal. See Figure 1 and Table 1 for details.
Models and Training
We used a previously published End-to-End Variational Network (E2E VarNet) with default hyper-parameters19. We trained separate models for acceleration rates of 2,4,6,8,10,12. We trained all models for 30 epochs.
Pathology Evaluation Procedure
We compared reconstructed images (RE) against corresponding annotated ground truth images (GT) at a slice-level as well as within the bounding box. We calculate the differences (MSE, NMSE) and similarity (SSIM, PNSR) between images as well as overall signal intensity mean and standard deviation. Metrics within labelled bounding boxes were compared against random patches of the same size within the same slice so as to avoid effects of patch size and background on the quantitative metrics. SSIM is calculated by obtaining a full SSIM map in the entire slice and then indexing annotated patches with bounding box information. SSIM is scaled by the max pixel value of the ground truth image with structural similarity package in skimage.
Results
Figure 3 shows the average value of MSE, NMSE, PSNR, and SSIM on the test set across acceleration rates. The MSE and NMSE increase more rapidly with increasing acceleration factor for abnormal bounding boxes (with pathology) vs. random bounding boxes (without pathology). The PSNR and SSIM are similar in low acceleration rates for the bounding boxes with pathology and without. However, the bounding boxes with pathology rapidly decrease with increasing acceleration factor after 6X. The normal brains (the second column) have better quantitative results than the abnormal brains in both slice-level images and annotated patches.Figure 4 shows results for three illustrative cases where three kinds of pathology (mass, edema, and craniectomy) were identified in separate images. We show the abnormal and random bounding boxes on reconstructions of 8X accelerated data. At high accelerations up to 8, we can see increase of NMSE and decrease of SSIM value in regions with pathology (example A). We also found large errors or low SSIM in image regions without labelled pathology (example B), especially at any edges of the image (example C). We found similar results in most kinds of pathology cases.
Conclusion
We evaluated deep learning-based reconstruction models in images with pathology at a range of acceleration factors. We designed an evaluation pipeline to quantify the differences between 1) pairs of abnormal vs. normal annotated patches in a slice and 2) pairs of reconstructed images in abnormal vs. normal brains. The quantitative analysis indicated that pathologies are potentially removed by deep learning-based algorithms trained by SSIM as loss function at a high acceleration rate. We also found the limit of using current metrics for evaluating loss in pathology for some cases where pathology is at the edge of the brain. In the future study, our quantitative assessment procedure can be used to design advanced loss functions and evaluate deep learning-based reconstruction algorithms in images with pathology.Acknowledgements
We would like to thank Mattew J. Muckley from Facebook AI Research for supporting this research.References
- Yang Y, Sun J, Li H, Xu Z. Deep ADMM-Net for compressive sensing MRI. In NeurIPS 2016 Dec 5 (pp. 10-18).
- Recht, M. P., Zbontar, J., Sodickson, D. K., Knoll, F., Yakubova, N., Sriram, A., ... & Zitnick, C. L. (2020). Using deep learning to accelerate knee MRI at 3 T: results of an interchangeability study. AJR. American journal of roentgenology, 215(6), 1421.
- Yang, G., Yu, S., Dong, H., Slabaugh, G., Dragotti, P. L., Ye, X., ... & Firmin, D. (2017). DAGAN: deep de-aliasing generative adversarial networks for fast compressed sensing MRI reconstruction. IEEE transactions on medical imaging, 37(6), 1310-1321.
- Knoll, F., Hammernik, K., Zhang, C., Moeller, S., Pock, T., Sodickson, D. K., & Akcakaya, M. (2019). Deep learning methods for parallel magnetic resonance image reconstruction. arXiv preprint arXiv:1904.01112.
- Shaul, R., David, I., Shitrit, O., & Riklin-Raviv, T. (2020). Subsampled brain MRI reconstruction by generative adversarial neural networks. Medical image analysis, 65, 101747 .
- Dar, S.U., Yurt, M., Shahdloo, M., Ildiz, M.E., Tinaz, B., & Çukur, T. (2020). Prior-Guided Image Reconstruction for Accelerated Multi-Contrast MRI via Generative Adversarial Networks. IEEE Journal of Selected Topics in Signal Processing, 14, 1072-1087.
- Pezzotti, N., Yousefi, S., Elmahdy, M.S., Gemert, J.V., Schulke, C., Doneva, M., Nielsen, T., Kastryulin, S., Lelieveldt, B.P., Osch, M.V., Weerdt, E.D., & Staring, M. (2020). An Adaptive Intelligence Algorithm for Undersampled Knee MRI Reconstruction. IEEE Access, 8, 204825-204838.
- Wang, S., Cheng, H., Ying, L., Xiao, T., Ke, Z., Liu, X., Zheng, H., & Liang, D. (2020). DeepcomplexMRI: Exploiting deep residual network for fast parallel MR imaging with complex convolution. Magnetic resonance imaging.
- Johnson, P.M., Muckley, M., Bruno, M., Kobler, E., Hammernik, K., Pock, T., & Knoll, F. (2019). Joint Multi-anatomy Training of a Variational Network for Reconstruction of Accelerated Magnetic Resonance Image Acquisitions. MLMIR@MICCAI.
- Weiss, T., Senouf, O., Vedula, S., Michailovich, O.V., Zibulevsky, M., & Bronstein, A.M. (2019). PILOT: Physics-Informed Learned Optimal Trajectories for Accelerated MRI. ArXiv, abs/1909.05773.
- Wang, Z., Bovik, A. C., Sheikh, H. R., & Simoncelli, E. P. (2004). Image quality assessment: from error visibility to structural similarity. IEEE transactions on image processing, 13(4), 600-612.
- Zhao, R., Zhang, Y., Yaman, B., Lungren, M. P., & Hansen, M. S. (2021). End-to-end AI-based MRI reconstruction and lesion detection pipeline for evaluation of deep learning image reconstruction. arXiv preprint arXiv:2109.11524.
- Muckley, M. J., Murrell, T., Radmanesh, A., Knoll, F., Huang, Z., Sriram, A., ... & Lui, Y. W. Properties of 2D MR image reconstructions with deep neural networks at high acceleration rates.
- Zbontar, J., Knoll, F., Sriram, A., Murrell, T., Huang, Z., Muckley, M. J., ... & Lui, Y. W. (2018). fastMRI: An open dataset and benchmarks for accelerated MRI. arXiv preprint arXiv:1811.08839.
- Knoll, F., Zbontar, J., Sriram, A., Muckley, M. J., Bruno, M., Defazio, A., ... & Lui, Y. W. (2020). fastMRI: A publicly available raw k-space and DICOM dataset of knee images for accelerated MR image reconstruction using machine learning. Radiology. Artificial intelligence, 2(1).
- Muckley, M. J., Riemenschneider, B., Radmanesh, A., Kim, S., Jeong, G., Ko, J., ... & Knoll, F. (2021). Results of the 2020 fastmri challenge for machine learning mr image reconstruction. IEEE transactions on medical imaging, 40(9), 2306-2317.
- Knoll, F., Murrell, T., Sriram, A., Yakubova, N., Zbontar, J., Rabbat, M., ... & Recht, M. P. (2020). Advancing machine learning for MR image reconstruction with an open competition: Overview of the 2019 fastMRI challenge. Magnetic resonance in medicine, 84(6), 3054-3070.
- Zhao, R., Yaman, B., Zhang, Y., Stewart, R., Dixon, A., Knoll, F., ... & Lungren, M. P. (2021). fastmri+: Clinical pathology annotations for knee and brain fully sampled multi-coil mri data. arXiv preprint arXiv:2109.03812.
- Sriram, A., Zbontar, J., Murrell, T., Defazio, A., Zitnick, C. L., Yakubova, N., ... & Johnson, P. (2020, October). End-to-end variational networks for accelerated MRI reconstruction. In International Conference on Medical Image Computing and Computer-Assisted Intervention (pp. 64-73). Springer, Cham.
Figures
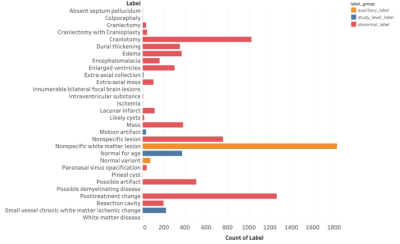
Figure 1: Summary statistics for categories of abnormalities annotated in fastMRI+. Of note, auxiliary labels categorized separately from abnormal labels were those slices identified to have ‘Nonspecific white matter lesion’ or ‘Normal variant’ as these were considered non-pathologic. All other labels were considered abnormal.
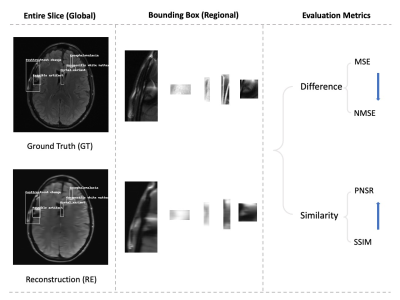
Figure 2: Diagram of the image quality evaluation pipeline. First, ground truth images (GT) and reconstructed images (RE) in different acceleration rates are labelled in three ways: the abnormal brains use fastMRI+ information; the random bounding boxes are created in the same abnormal brains; the normal brains are labelled similar abnormal brains. Second, annotated patches are extracted from the entire slice. Each pair of GT and RE is used in the last evaluation procedure for calculating the difference (MSE and NMSE) and similarity (PSNR, SSIM).
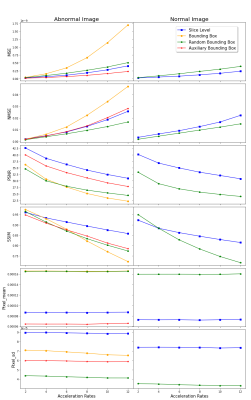
Figure 3: Mean evaluation metrics across accelerations for normal (right) and abnormal (left) image content. In calculating the green lines, all slices with too large bounding boxes to avoid overlapping are removed.
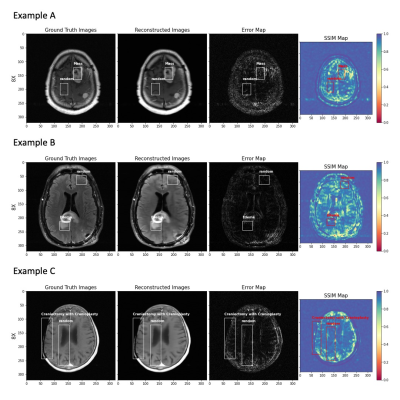
Figure 4: fastMRI+ labels of pathology and generated random bounding boxes of same dimension in three illustrative cases at 8X: (A) enhancing intracranial masses, (B) edema, and (C) Craniectomy with Cranioplasty, showing from left to right: ground truth image, reconstructed image, error map, and SSIM map. The random bounding boxes avoid overlap with label(s) and are required to be at least 80% within the masked region of the head.
DOI: https://doi.org/10.58530/2023/3097