3096
Geometric Constrained Deep Learning for Motion Correction of Fetal Brain MR Images
Laifa Ma1,2, Liangjun Chen1, Fenqiang Zhao1, Zhengwang Wu1, Li Wang1, Weili Lin1, He Zhang3, Kenli Lin2, and Gang Li1
1University of North Carolina at Chapel Hill, Chapel Hill, NC, United States, 2Hunan University, Changsha, China, 3Fudan University, Shanghai, China
1University of North Carolina at Chapel Hill, Chapel Hill, NC, United States, 2Hunan University, Changsha, China, 3Fudan University, Shanghai, China
Synopsis
Keywords: Image Reconstruction, Brain
Robust motion correction of fetal brain MRI slices is crucial for fetal brain volume reconstruction. However, conventional methods can only handle a limited range of motion. Hence, a deep learning model based on prior geometric constraints is proposed to predict the motion of 2D slices. It consists of a global and a relative motion estimation network. Sharing features between two networks make the model to learn more unique feature representations for global motion correction. Moreover, we present a control point-based approach to simulate complex fetal motion trajectories. The experimental results demonstrate that the proposed method is effective and efficient.Introduction
Fetal brain magnetic resonance imaging (MRI) is becoming increasingly important in prenatal screening[1].However, the unpredictable motion of both the fetus and the mother makes obtaining high-resolution fetal 3D MR images extremely challenging[2]. To overcome this challenge, fast imaging methods[3] are typically used to acquire thick and motion-corrupted 2D stacks in different orientations. Then, iterative operations of motion correction and super-resolution reconstruction are used to estimate fetal brain high-resolution 3D MR images[2]. Although the traditional iterative motion correction and reconstruction approaches have proven effectiveness, they have two major limitations: 1) failures usually occur in estimating large motion; and 2) existing methods heavily rely on good initialization of transformation parameters. Hence, we propose a geometric constraint-based deep learning model (GCDL) to predict the arbitrary motion of fetal brain MRI slices in a standard anatomical space. GCDL consists of a global motion estimation network and a local relative motion estimation network. The learned features by the relative motion estimation network are further shared with the global motion network to learn more unique feature representations for the global motion correction, thus improving the performance.Methods and Materials
Our method consists of a global motion regression network and a relative motion regression network, as shown in Fig. 1. The goal of our method is to accurately predict fetal brain motion in a standard space. For this purpose, the relative motion between two adjacent slices (Sn, Sn-1) in a stack is exploited as a geometric constraint to limit the search space. Specifically, a relative motion network is proposed to estimate the relative motion between slices. Then, the learned features are further shared to the global motion network to learn more unique feature representations for global motion correction, and a weight-learnable strategy is employed to balance the contributions of global and local networks in the model. Sharing features between the two networks enables competitive and cooperative action. This approach thus provides geometric constraints for predicting the absolute motion of the fetus, reduces the search space, and enables the model to achieve better performance.To train the prediction model, we synthesized a large number of fetal brain MRI stacks with a wide variety of slice thickness and fetal motion levels using 3D fetal brain MRI data. To simulate the speed of fetal movement, a control point-based approach was developed. In detail, by utilizing the cubic spline interpolation between randomly generated control points, smooth fetal motion trajectories can be generated. Different numbers of control points were applied to simulate the movement speed of the fetus. For instance, when there are fewer control points, the generated motion trajectory is relatively smooth, and the movement speed between fetal slices is relatively slow. In comparison, the generated motion trajectory with increased number of control points is more complex, suggesting vigorous moving. Fig. 2 shows 6 Degrees of Freedom motion trajectories of the fetus generated by different control points.
We developed the model based on 279 T2-weighted fetal brain MRI volumes between 21 and 38 weeks of gestational age. The fetal volumes were reconstructed using NiftyMIC[4], and then aligned onto a fetal brain atlas with 0.8 mm isotropic resolution. The data sets applied to simulate the fetal brain motion were divided into training (194 volumes), validation (30), and test (55 volumes) sets. The 2D slices were simulated with 0.8 mm × 0.8 mm, slice thickness is 4 mm. The number of control points used to generate fetal motion trajectories at different speeds are 4, 5, 6, and 7. For each volume and different number of control points, we generated 400 fetal motion trajectories in coronal, axial and sagittal orientations, respectively. we respectively set rotation angles of slices from -40° to 40°, translation parameters were set to -12 mm to 12. The Cartesian coordinate anchor points were used to parameterize the motion of fetuses, where three non-collinear points are used to determine the position of a slice in standard space.
Results
The standard metrics: Cross Correlation (CC), Structural Similarity (SSIM), and Peak Signal-to-Noise Ratio (PSNR), are used to evaluate the performance of methods. On coronal, axial, and sagittal orientations, the average values achieved by the proposed method are 0.851, 18.796, and 0.694 according to CC, PSNR, and SSIM, respectively. Our method achieved superior performance against SVRnet[5], which only exploits the intensity information of the single slice. Fig. 3 presents a visual comparison between the typical slices of ground truth, the slices obtained by GCDL, and the slices obtained by SVRnet, demonstrating the outstanding performance obtained by the proposed method.Conclusions
In this work, to achieve accurate fetal brain MRI motion correction, a deep learning model based on geometric constraints is proposed to predict the arbitrary motion of fetal brain MRI slices. Leveraging the geometric information, the proposed method can automatically learn the correlation between slices, thus leading to superior performance. Moreover, we present a control point-based approach to simulate fetal motion trajectories. The experimental results on simulated fetal brain stacks demonstrate that the proposed method is effective and efficient.Acknowledgements
This work was partially supported by NIH grants (MH116225, MH117943 and MH123202 to G.L.).Figures
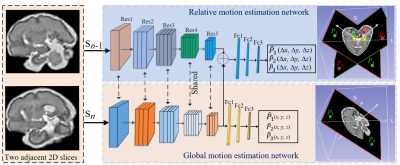
Fig. 1. An overview of the proposed GCDL for anchor points regression, including a global motion estimation network (GMEnet) and a relative motion estimation network (RMEnet). RMEnet is proposed to learn the geometric information between slices. The features learned by RMEnet are shared to GMEnet to learn more unique feature representations for global motion correction.
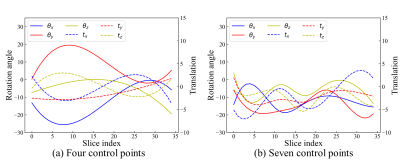
Fig. 2. Illustration of 6 Degrees of Freedom motion trajectories of the fetus generated by four and seven control points in standard space. Each fetal motion trajectory corresponds to a fetal stack of slices. Obviously, movements between fetal brain MRI slices simulated with seven control points (b) are more sophisticated than those generated with four control points (a).
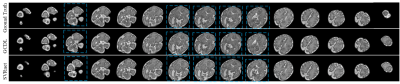
Fig. 3. The visualization of ground truth, and results obtained by GCDL and SVRnet on a randomly selected 32-week stack. The slices marked by the blue dashed box, which demonstrate the outstanding performance obtained by the proposed method.
DOI: https://doi.org/10.58530/2023/3096