3083
Accelerated whole-heart MRI for congenital heart disease patients using a motion-corrected deep learning reconstruction network
Andrew Phair1, Anastasia Fotaki1, Lina Felsner1, Haikun Qi2, René M. Botnar1, and Claudia Prieto1
1School of Biomedical Engineering and Imaging Sciences, King's College London, London, United Kingdom, 2School of Biomedical Engineering, ShanghaiTech University, Shanghai, China
1School of Biomedical Engineering and Imaging Sciences, King's College London, London, United Kingdom, 2School of Biomedical Engineering, ShanghaiTech University, Shanghai, China
Synopsis
Keywords: Image Reconstruction, Cardiovascular
A deep learning reconstruction framework, trained in an end-to-end fashion and incorporating both a non-rigid respiratory motion estimation network and a motion-informed model-based reconstruction network, has been previously demonstrated to enable good quality images from seven-fold undersampled acquisitions for coronary magnetic resonance angiography applications. Herein, we apply the framework to whole-heart MRI scans of patients with congenital heart disease, enabling fast reconstruction of 7×-accelerated acquisitions and achieving image quality comparable to that of state-of-the-art patch-based low-rank iterative techniques.Introduction
Cardiovascular magnetic resonance angiography (CMRA) is established for anatomical assessment in patients with congenital heart disease (CHD)1. Conventional acquisition strategies rely on diaphragmatic respiratory gating (dNAV T2prep-bSSFP), leading to long and unpredictable scan times and residual motion artefacts. Furthermore, achieving scan acceleration via undersampling k-space often involves the use of iterative reconstruction methods with long reconstruction times.Recently, a deep learning reconstruction framework incorporating a motion estimation network and a motion-informed model-based reconstruction network, trained in an end-to-end manner, has been proposed for free-breathing whole-heart coronary magnetic resonance angiography with 100% respiratory scan efficiency2. In that study, a combination of fully sampled and two-to-three-fold undersampled data was utilised to train the network, which was then successfully applied to reconstruct images from 7×-undersampled acquisitions (a ~2.5-minute scan). Herein, we propose to apply this reconstruction framework to 7×-undersampled 3D whole-heart scans of CHD patients to enable 3D whole-heart acquisition in 1-2 minutes.
Methods
40 adult patients with CHD were scanned on a 1.5 T system (MAGNETOM Aera, Siemens Healthcare) using an ECG-triggered free-breathing T2-prepared bSSFP sequence3 and the following imaging parameters: FOV = 400 mm × 300 mm × 72-108 mm, resolution = 1.5 mm × 1.5 mm × 1.5 mm, flip angle = 90°, T2-prep duration = 40 ms, TE = 1.75 ms, TR = 238ms, coronal orientation. For each patient, two scans were acquired: one with three-or-four-fold undersampling and another with seven-fold undersampling. A 2D image navigator (iNAV) was acquired during each heartbeat for respiratory binning, and to ensure each respiratory bin exhibited incoherent undersampling artefacts a variable-density Cartesian trajectory with spiral-like sampling (VD-CASPR)4 was utilised.The data were randomly sorted into a training set (25 patients) and a testing set (15 patients). The training data (3-4× undersampled) were used to generate a ground truth and zero-filled image for each of four respiratory bins, as shown in Figure 1 (a). Since fully sampled data sets were not obtained, the ground truth images were reconstructed using the iterative patch-based low-rank method 3D-PROST3.
The deep learning reconstruction network (MoCo-MoDL)2 consisted of a diffeomorphic motion estimation network5, which estimated the motion fields between respiratory bins, and a motion-informed model-based reconstruction network, which enforced data consistency between the reconstruction and the input zero-filled respiratory-bin images. A schematic of the network structure is shown in Figure 2. The network was trained on the 25-patient training set over 200 epochs and then tested on 7× prospectively undersampled scans of 15 patients in the manner depicted in Figure 1 (b). The output of the network was a motion-corrected whole-heart image in the reference respiratory phase.
Results
Average acquisition times were 1.5±0.3 minutes (7×-undersampled) and 3.2±0.7 minutes (3-4×-undersampled). Coronal cardiac images obtained using the MoCo-MoDL network are presented for four representative patients in Figure 3. In each case, the input to the network (a 7×-undersampled zero-filled soft-gated respiratory-bin image) is shown for comparison. Additionally, a “ground truth” PROST reconstruction calculated from the 3-4× undersampled scan of the same patient is presented. We note that while these ground truth reconstructions are generated following the same steps as depicted in Figure 1 (a) for the training datasets, they are not included in the network training and are only calculated for presentation.Discussion
Visual inspection of the image quality in Figure 3 suggests that the MoCo-MoDL network achieves comparable image quality to the 3D-PROST “ground truth” images despite the higher undersampling factor. Additionally, the network reconstruction avoids the long reconstruction time associated with iterative algorithms such as 3D-PROST. However, we note that the images produced by the network tend to not be as sharp as those from the 3D-PROST reconstructions with a lower undersampling factor.Future work will focus on tuning the hyperparameters of the network and incorporating non-rigid motion correction into the ground truth 3D-PROST reconstruction.
Conclusion
Fast reconstruction of 7×-undersampled whole-heart scans of CHD patients was achieved by utilising an end-to-end trained deep learning network to simultaneously estimate non-rigid respiratory motion fields and implement these fields in a motion-corrected model-based reconstruction.Acknowledgements
This work was supported by the following grants: (1) EPSRC P/V044087/1; (2) BHF programme grant RG/20/1/34802, (3) Wellcome/EPSRC Centre for Medical Engineering (WT 203148/Z/16/Z), (4) Millennium Institute for Intelligent Healthcare Engineering ICN2021_004, (5) FONDECYT 1210637 and 1210638, (6) IMPACT, Center of Interventional Medicine for Precision and Advanced Cellular Therapy, ANID FB210024.References
- Fratz, S., Chung, T., Greil, G. F., Samyn, M. M., Taylor, A. M., Valsangiacomo Buechel, E. R., Yoo, S. & Powell, A. J. (2013). Guidelines and protocols for cardiovascular magnetic resonance in children and adults with congenital heart disease: SCMR expert consensus group on congenital heart disease. Journal of Cardiovascular Magnetic Resonance, 15(1), 1-26.
- Qi, H., Hajhosseiny, R., Cruz, G., Kuestner, T., Kunze, K., Neji, R., Bonar, R. M. & Prieto, C. (2021). End‐to‐end deep learning nonrigid motion‐corrected reconstruction for highly accelerated free‐breathing coronary MRA. Magnetic Resonance in Medicine, 86(4), 1983-1996.
- Bustin, A., Ginami, G., Cruz, G., Correia, T., Ismail, T. F., Rashid, I., Neji, R., Botnar R. M. & Prieto, C. (2019). Five‐minute whole‐heart coronary MRA with sub‐millimeter isotropic resolution, 100% respiratory scan efficiency, and 3D‐PROST reconstruction. Magnetic Resonance in Medicine, 81(1), 102-115.
- Prieto, C., Doneva, M., Usman, M., Henningsson, M., Greil, G., Schaeffter, T., & Botnar, R. M. (2015). Highly efficient respiratory motion compensated free‐breathing coronary MRA using golden‐step Cartesian acquisition. Journal of Magnetic Resonance Imaging, 41(3), 738-746.
- Munoz, C., Qi, H., Cruz, G., Küstner, T., Botnar, R. M., & Prieto, C. (2022). Self-supervised learning-based diffeomorphic non-rigid motion estimation for fast motion-compensated coronary MR angiography. Magnetic Resonance Imaging, 85, 10-18.
Figures
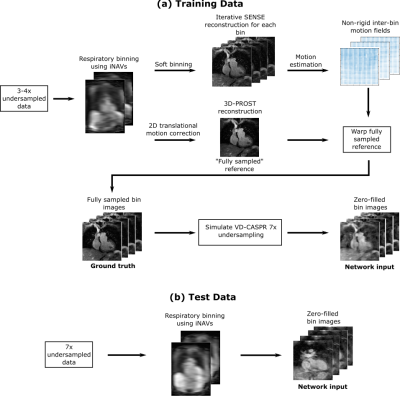
Figure 1. Schematic of the data processing pipeline. (a)
The training data is sorted into respiratory bins and an iterative SENSE reconstruction
is performed for each bin. A “fully sampled” reference image is obtained via 3D-PROST and then warped by inter-bin motion fields estimated
from the SENSE reconstructions. The resultant bin images are considered to be
the ground truth. k-Space undersampling with a VD-CASPR trajectory is used to
obtain zero-filled bin images. (b) The test data is sorted into respiratory
bins and a zero-filled reconstruction obtained for each bin.

Figure 2. Schematic of the
deep learning framework which consists of a respiratory non-rigid motion
estimation (ME) network and a motion-corrected model-based reconstruction
network (MoDL) trained simultaneously in an end-to-end manner.

Figure 3. Zero-filled respiratory-bin images from the
7×-undersampled acquisitions (left), “ground truth” iterative 3D-PROST
reconstructions from 3-4×-undersampled acquisitions (centre) and the MoCo-MoDL
network reconstructions from the 7×-undersampled input (right) for four
patients in the test set.
DOI: https://doi.org/10.58530/2023/3083