3067
Automated Intracranial Artery Labeling in Patients with Cerebrovascular Steno-occlusive Diseases
Lixin Liu1, Yi Lv2, Peirong Jiang3, He Wang1,4,5, and Zhensen Chen1,5
1Institute of Science and Technology for Brain-Inspired Intelligence,Fudan University, Shanghai, China, 2School of Compute Science and Technology, Beijing Institute of Technology, Beijing, China, 3Department of Radiology, Fujian Medical University Union Hospital, Fuzhou, China, 4Human Phenome Institute, Fudan University, Shanghai, China, 5Key Laboratory of Computational Neuroscience and Brain-Inspired Intelligence (Fudan University), Ministry of Education, Shanghai, China
1Institute of Science and Technology for Brain-Inspired Intelligence,Fudan University, Shanghai, China, 2School of Compute Science and Technology, Beijing Institute of Technology, Beijing, China, 3Department of Radiology, Fujian Medical University Union Hospital, Fuzhou, China, 4Human Phenome Institute, Fudan University, Shanghai, China, 5Key Laboratory of Computational Neuroscience and Brain-Inspired Intelligence (Fudan University), Ministry of Education, Shanghai, China
Synopsis
Keywords: Data Processing, Blood vessels
Labeling of intracranial arteries is important for computer-aided diagnosis of cerebrovascular diseases and quantitative analysis of intracranial vasculature. Performance of the previously proposed automated intracranial artery labeling method based on Graph Neural Network (GNN) is limited in datasets with overt cerebrovascular steno-occlusive diseases. In this study, we improved the generalizability of the GNN-based method by using dedicated data augmentation and spatial normalization strategy. The results show that our method is more robust than the previous method in ischemic stroke patients with overt intracranial stenosis or occlusion.Introduction
Labeling of intracranial arteries is of great value for computer-aided diagnosis of cerebrovascular diseases and quantitative analysis of intracranial vasculature1. Chen et al.2 proposed an automated intracranial artery labeling method based on Graph Neural Network (GNN). Although excellent performance was achieved, this method is limited in its generalizability, because the GNN model was trained on datasets without overt cerebrovascular steno-occlusive diseases and relied on an accurate segmentation of cerebral vessels. In this study, we aimed to improve the generalizability of the GNN-based automated intracranial artery labeling method for patients with overt cerebrovascular steno-occlusive diseases.Methods
GNN modelIn Chen et al.2, intracranial vascular network was represented by a graph with nodes and edges. They used the message passing GNN framework, where input features are encoded in an encoder layer and restored with additional labeling features in a decoder layer. Five datasets of labeled intracranial artery centerlines were obtained by using the iCafe tool3, of which 70% (N=512) were used for training, 15%(N=108) for validation and 15%(N=108) for testing. Majority of the subjects in these datasets had a normal and complete intracranial vasculature.
In this study, we made improvements to Chen et al’s method from two aspects, i.e. data augmentation and spatial normalization.
Data Augmentation: To mimic the missing of some vessel segments in brain with overt cerebrovascular steno-occlusive diseases, we performed random removal of the proximal large vessels (e.g. ICA, A1, M1 and P1) on the above-mentioned five datasets. In addition, since the branch number and length of distal arteries may vary over different subjects, imaging protocols, MRI scanner, data collection sites and vessel segmentation methods, we also randomly removed some branches and reduced length of distal arteries (i.e. M2, A2, and P2).
Spatial Normalization: Given that the coordinates of the same type of vessel may differ over datasets due to inconsistency in position planning of TOF scans, which may pose a challenge to the GNN model, we proposed to normalize spatial coordinates of the data before retraining GNN model, i.e. shift origin of the coordinate system to the center of CoW. In this study, the center of CoW was calculated as the average location of the seven vessel segments that make up the CoW (i.e. anterior communicating artery (AcomA), right and left posterior communicating artery (Pcomm), right and left A1, right and left P1) from the ground truth.
Datasets
We retrained the GNN model on the above-mentioned 5 datasets (https://github.com/clatfd/GNN-ART-LABEL/tree/master/graph/graphsim) after data augmentation and spatial normalization, of which also 70% (N=973) were used for training, 15%(N=209) for validation and 15%(N=208) for testing.
To compare Chen’s method and ours, three datasets were used for testing. Testing Dataset 1 included 15 cases that were randomly selected from the previous five datasets. Testing Dataset 2 were the Testing Dataset 1 with some vessel segments randomly removed. Testing Dataset 3 is a private dataset and includes 15 ischemic stroke patients with overt intracranial stenosis or occlusion. Cerebral arteries in Testing Dataset 3 were semi-automatically extracted from 3D TOF-MRA images (field of view: 200 × 192 × 106 mm3, spatial resolution: 0.52 × 0.72 × 1.2 mm3) using 3D Slicer.
Evaluation Metrics
We used the accuracy of node labeling (Node_Acc) and edge labeling (Edge_Acc) as evaluation metrics. The detection accuracy for 7 major bifurcation types (ICA-OA, ICA-M1-A1, ICA-Pcomm, A1-AcomA-A2, M1-M2, VA-P1, P1-Pcomm-P2) were also calculated.
Results and Discussion
The labeling accuracy of the two methods for nodes and edges as well as the major bifurcations are shown in Table 1 and 2. Example cases from the 3 testing datasets are shown in Fig. 1. As expected, both methods show a large decrease of labeling accuracy in Test Dataset 2 and Dataset 3 compared to the Testing Dataset 1, suggesting that the missing of some intracranial arteries or decreased visibility of distal arteries under overt cerebrovascular steno-occlusive diseases posed a large challenge to the GNN model. For Testing Dataset 1, results of the two methods are similar, indicating that our method is applicable for data without missing vessel segments. Importantly, our method demonstrates a greater robustness for abnormal intracranial vasculature, with an edge accuracy of 60.73% and 60.28% for Testing Dataset 2 and Testing Dataset 3, respectively. Note that in real application, the labeling of edge may be more useful than labeling of the nodes, and the two types of labeling could be further combined with a heuristic strategy as in Chen et al2.Conclusion
We have successfully demonstrated that by using data augmentation and spatial normalization strategy, robustness of the GNN-based intracranial artery labeling method in patients with overt cerebrovascular steno-occlusive diseases could be improved. However, to make the method usable in clinical practice, further improvement of the labeling accuracy is needed.Acknowledgements
This study was supported in part by the Shanghai Natural Science Foundation (No. 22ZR1403900) and Fujian Provincial Science and Technology Guiding Project (2021Y0017).References
1. Robben D, Türetken E, Sunaert S, Thijs V, Wilms G, Fua P, Maes F, Suetens P. Simultaneous segmentation and anatomical labeling of the cerebral vasculature. Med Image Anal. 2016 Aug; 32:201-15.2. Li Chen, Thomas Hatsukami, Jenq-Neng Hwang, Chun Yuan, “Automated intracranial artery labeling using a graph neural network and hierarchical refinement,” in International Conference on Medical Image Computing and Computer-Assisted Intervention. Springer, 2020, pp. 76–85.
3. Chen L, Mossa-Basha M, Balu N, Canton G, Sun J, Pimentel K, Hatsukami TS, Hwang JN, Yuan C. Development of a quantitative intracranial vascular features extraction tool on 3D MRA using semiautomated open-curve active contour vessel tracing. Magn Reson Med. 2018 Jun;79(6):3229-3238.
Figures
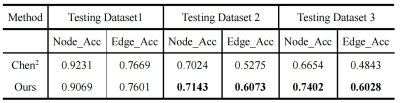
Table 1. The labeling accuracy of the two methods for nodes and edges.
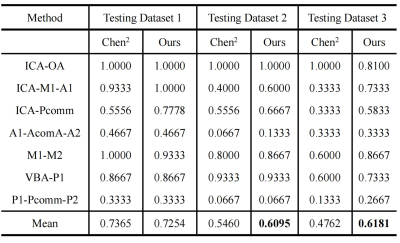
Table 2. The predictions accuracy of the 7 major bifurcations by the two methods.
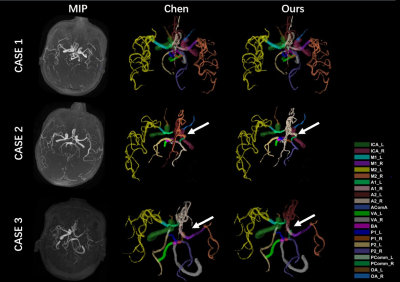
Figure 1. Arterial labeling results for the three example cases from the 3 testing datasets. In CASE 1 (Test Dataset 1), the vessels are not missing and both methods performed well. In CASE 2 (Test Dataset 2), the right M1 and M2 were artificially removed. CASE 3 (Test Dataset 3) misses the right ICA. On these two cases, our method predicts 13 and 11 vessels correctly, while Chen's method predicts 10 and 9 vessels correctly, indicating that our method is more robust.
DOI: https://doi.org/10.58530/2023/3067